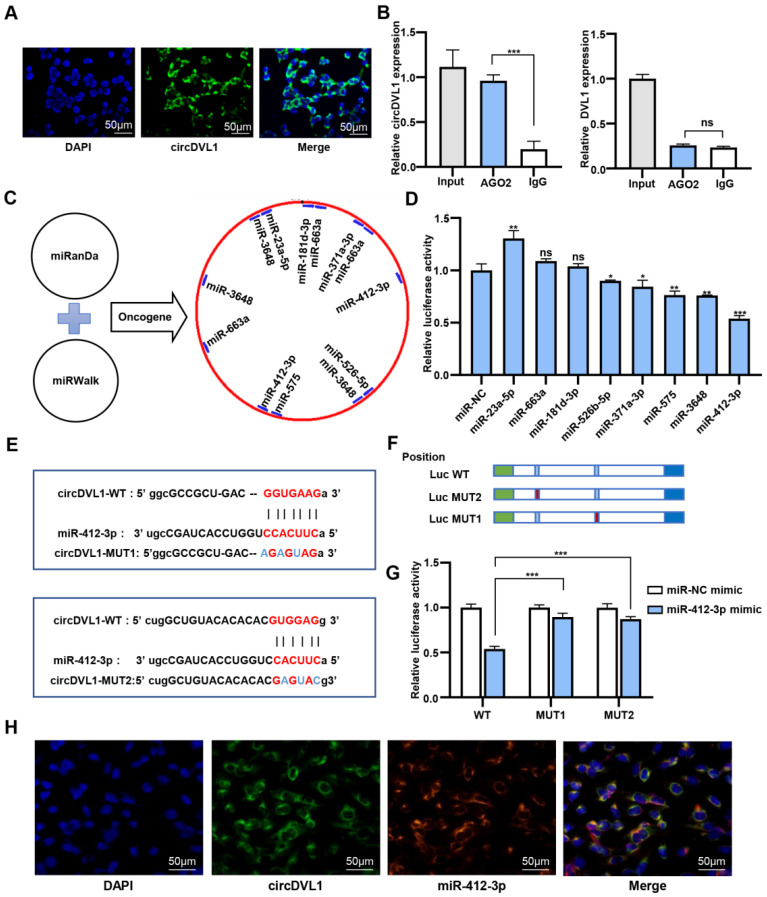
Figure 4.
CircDVL1 functions as a molecular sponge for miR-412-3p in ccRCC cells. (A) FISH analysis showed that circDVL1 (green) and DAPI (blue) localized to the cytoplasm of 786-O. Scale bar: 50 µm. (B) RNA immunoprecipitation (RIP) and RT-qPCR assays to detect circDVL1 and DVL1 were immunoprecipitated by AGO2 and IgG antibodies in 786-O. (C) The schematic flowchart shows the pipelines that can detect oncogenic miRNA bound to circ-DVL1 3'-UTR via the online bioinformatics network. (D) Dual-luciferase reporter assays were used to evaluate candidate miRNAs associated with circDVL1 in 293T cells. (E and F) Sequence of Wild-type (WT) and mutated (MUT) putative miR-412-3p-binding sites in the 3'-UTR of circDVL1. (G) Relative luciferase activities were examined in 293T after transfection with circDVL1-WT or circDVL1-MUT and miR-412-3p mimic or miR-NC. (H) RNA CO-FISH assay for circDVL1 (green), miR-412-3p (red), DAPI (blue), and purple in merge in 786-O cells. miR-NC mimic was used as the miRNA mimic negative control. Scale bar, 100 µm. Data are presented as mean ± SD. *P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001, no significant (NS).