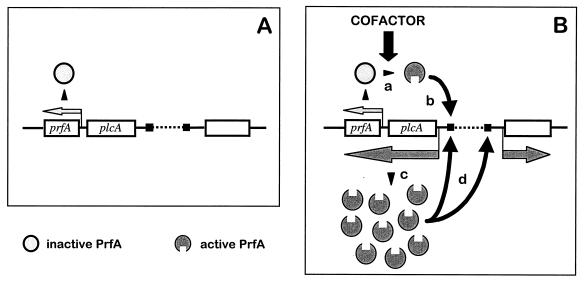
FIG. 13.
Model for the mechanism of PrfA-mediated regulation. This model predicts that PrfA can undergo an allosteric transition between inactive (light gray) and active (dark gray) forms upon interaction with a low-molecular-weight hypothetical cofactor (see text for details). (A) Resting PrfA system. There is no cofactor, and the PrfA protein is synthesized at low, basal levels from the monocistronic transcripts generated from growth phase-regulated promoters in front of the prfA gene (small light gray arrow). (B) Activated PrfA system. If L. monocytogenes senses a suitable combination of environmental signals (i.e., 37°C and cytoplasmic environment), the intracellular concentration of the cofactor increases, leading to activation of the PrfA protein (a), which binds with increased affinity to the PrfA-boxes (black squares) in PrfA-regulated promoters (b); the transcriptionally active PrfA form causes the synthesis of more PrfA (in active conformation) via the positive autoregulatory loop generated by the PrfA-dependent bicistronic plcA-prfA transcript (c), thereby inducing the transcription of all PrfA-dependent genes (d) (dark gray arrows represent PrfA-dependent transcripts; the empty rectangle on the right represents any PrfA-dependent gene). The PrfA regulon remains switched on as long as the cofactor is present in the bacterial cytoplasm, but is rapidly switched off if the activating environmental signals cease and the concentration of the cofactor drops. (Reproduced from reference 673 with permission of the publisher.)