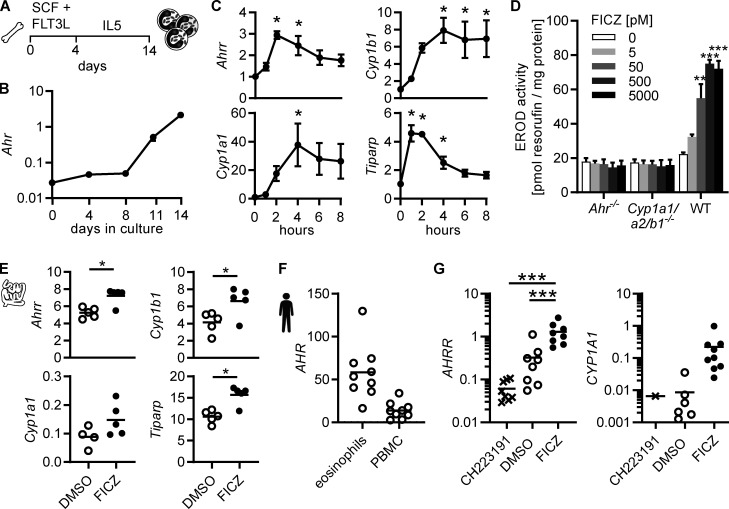
Figure 3.
Eosinophils activate the canonical AHR pathway in response to AHR ligands. (A) Schematic of BMDEo cultures. (B) Gene expression in BMDEo at different timepoints in culture was determined by qPCR and normalized to Hprt. Shown is the mean ± SEM of three independent experiments. (C) BMDEo were cultured to day 14 and treated with 5 nM FICZ or DMSO for indicated times. qPCR of AHR target genes was normalized to Hprt and DMSO-treated controls. Data are mean ± SEM of n = 4 independent experiments. *, P < 0.05, one-way ANOVA to DMSO control. (D) BMDEo of different genotypes were cultured to day 14 and treated with different concentrations of FICZ or DMSO control for 4 h before ethoxyresorufin-O-deethylase activity was measured. Data are mean ± SEM of n = three independent experiments. **, P < 0.01; ***, P < 0.001, one-way ANOVA to DMSO control. (E) Small intestinal eosinophils were FACS-sorted and cultured in vitro in the presence of 5 nM FICZ or DMSO control and 2 ng/ml IL-5 for 2 h. Gene expression was determined by qPCR and normalized to Hprt. *, P < 0.05, paired t test. Data are representative of two independent experiments with n = 4–5 mice. (F and G) Human eosinophils and PBMCs were isolated from blood of normal donors. Gene expression was analyzed by qPCR and normalized to HPRT. (F) Eosinophils and PBMCs were cultured for 4 h with DMSO. (G) Eosinophils were cultured with DMSO, 3 μm CH223191, or 5 nM FICZ for 4 h. CYP1A1 expression was detectable in only one of nine CH223191-treated and in six of nine DMSO-treated samples. Data are from nine donors. ***, P < 0.001, repeated-measure one-way ANOVA.