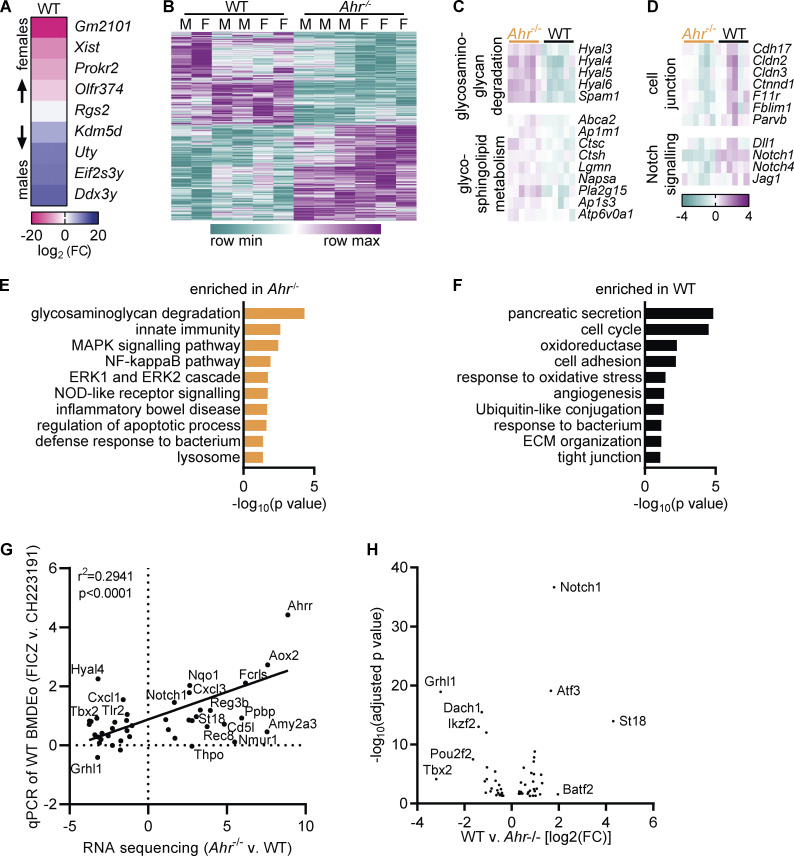
Figure S3.
AHR deficiency alters the transcriptome of intestinal eosinophils. (A) Differentially expressed genes between eosinophils from the small intestine of male and female WT mice with an adjusted P value <0.1. (B) Heatmap of differentially expressed genes (adjusted P value <0.05) between WT and Ahr–/– eosinophils from the small intestine. (C and D) Examples of functionally defined gene subsets from the leading edge of the analysis in Fig. 4 D. Shown is the log2(FC) to the geometric mean of TPM + 1. (E and F) Enriched pathways (P < 0.1) identified by DAVID based on differentially expressed genes between WT and Ahr–/– eosinophils (|FC| > 2, adjusted P value <0.05). (G) From the RNA sequencing dataset of small intestinal eosinophils, 55 genes with differential expression between WT and Ahr–/– were manually selected and analyzed by qPCR in BMDEo. Of those, 11 genes were undetectable in BMDEo. Correlation of RNA sequencing results with qPCR results is shown for WT BMDEo treated for 4 h with 5 nM FICZ versus 3 μM CH223191. Shown is log2(FC). qPCR data are from one experiment conducted in technical triplicate. (H) Volcano plot of transcription factors with differential expression between WT and Ahr–/– eosinophils.