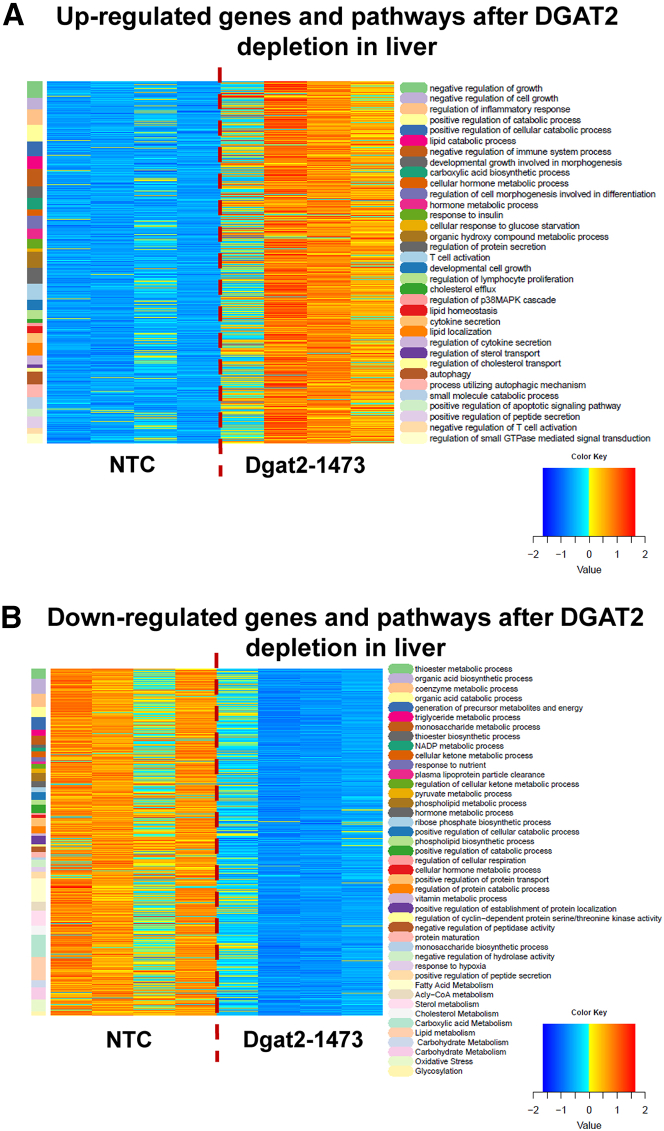
Figure 5.
Dgat2 silencing by Dgat2-1473 elicits significant changes in many pathways and genes in the livers of genetically obese mice with NASH
DEBrowser data that came from the DolphinNext RNA-seq pipeline (Figure 5) was first filtered to eliminate genes whose expression level was not above 10 in any sample, and then DESeq2 was employed to determine DE genes, using an adjusted p value of 0.05 as the cutoff and requiring at least a 1.5-fold change, up or down. The list of DE genes was then analyzed using the enrichGO function in the clusterProfiler package. The pathways were simplified using its simplify function with options of p value cutoff of 0.05. This was followed by manually specified merging of similar pathways to produce heatmaps. DE genes were clustered according to the pathways they were involved in, and the pathways (color coded) were displayed on the left column alongside the heatmap. Color scheme for the DE gene presentation was produced by Z-scoring method (−2 to +2 standard deviation from the mean) on expression levels normalized for sample depth by DESeq2. Heatmap representation of (A) upregulated and (B) downregulated DE genes clustered by the pathways in which they are involved.