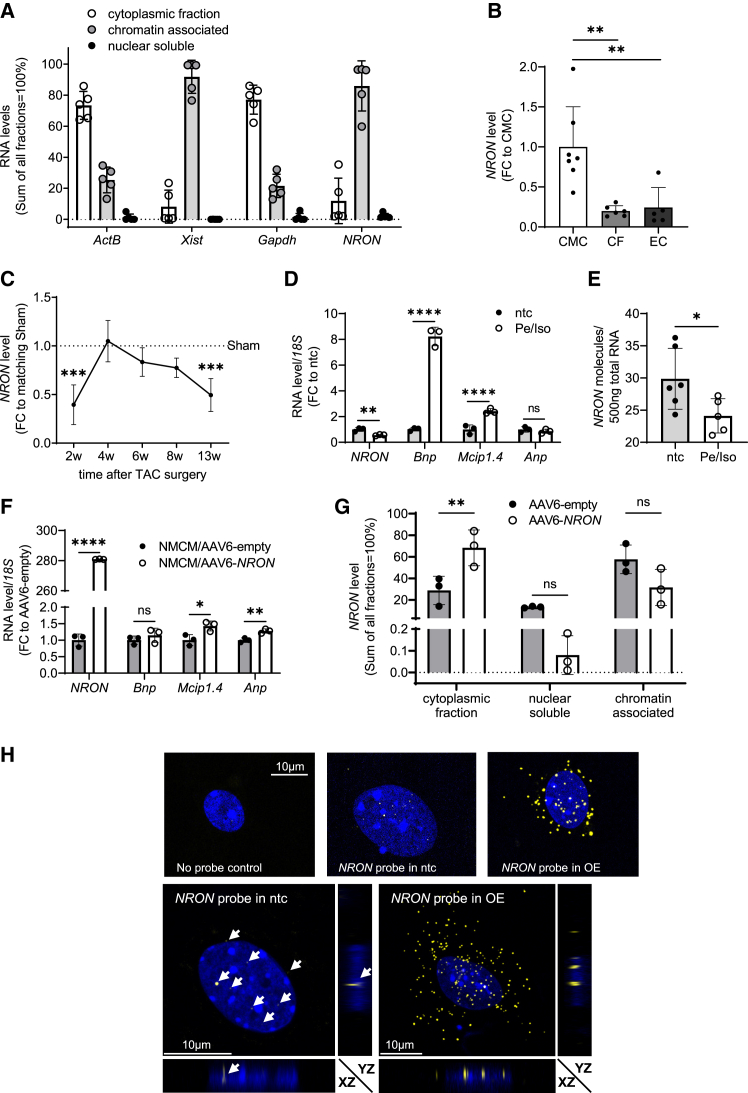
Figure 1.
NRON level in neonatal mouse cardiomyocytes and heart fractions and after transaortic constriction
(A) Subcellular distribution of NRON in NMCM subfractions. Gapdh, ActB, and Xist mRNA levels were measured as positive controls. Data are calculated as fold change (FC) to the main fraction of the transcript measured by qPCR (mean ± SD; n = 5). (B) Distribution of NRON in murine heart fractions: cardiomyocytes (CMCs), cardiac fibroblasts (CFs), and endothelial cells (ECs). The expression level was measured by qPCR, and ActB was used as a reference gene. Data are calculated as FC to the main fraction (CMCs) of the transcript (mean ± SD; n = 5–7). ∗∗p < 0.01; one-way Anova with Tukey's multiple comparisons test. (C) The expression of NRON in the whole mouse heart after 2, 4, 6, 8, and 13 weeks of TAC surgery. The expression level was measured by qPCR, and ActB was used as a reference gene. Data are calculated relative to Sham samples of each time point (mean ± SD; n = 5–8). ∗∗∗p < 0.001; one-way ANOVA with Sidak multiple comparisons test. (D) RNA levels in neonatal mouse cardiomyocytes (NMCMs) after treatment with 100 μM Pe/Iso for 48 h. The RNA level was measured by qPCR relative to 18S RNA. The FC was calculated using the respective RNA levels in not-treated controls (ntcs). All data are mean ± SD; n = 3. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; Student's t test. (E) Absolute quantification of NRON molecules in NMCMs. A plasmid standard containing murine NRON was used for quantification, and the level of NRON was measured by qPCR. NMCMs were either not treated (ntcs) or treated with 100 μM Pe/Iso for 48 h. n = 6 technical replicates. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; Student's t test. (F) RNA levels in NMCMs after the transduction of AAV6-NRON. The RNA level was measured by qPCR relative to 18S RNA. The FC was calculated using the respective RNA level in AAV6-empty-treated cells as control. All data are mean ± SD; n = 3. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; Student's t test. (G) Subcellular distribution of NRON in NMCM subfractions of cells treated with 5 × 103 AAV6-empty or AAV6-NRON viral particles. Data are calculated by taking the sum of all fractions as 100% (mean ± SD; n = 3). The RNA level was measured by qPCR. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; two-way ANOVA with Tukey's multiple comparisons test. (H) Staining of NRON in NMCMs using RNA-FISH. (Top panel) RNA-FISH of a single nucleus from untreated NMCMs stained with no probe as control and NMCMs infected with AAV6-empty (ntcs) or AAV6-NRON (overexpressed [OE]) viral particles stained with probe for NRON. (Bottom panel) Orthogonal sections from z stack confocal images of NMCMs infected with AAV6-empty (ntcs) or AAV6-NRON (OE) viral particles and stained with RNA-FISH. DAPI was used to stain the nucleus, and NRON was stained using an Alexa Fluor 546 tagged probe set. Representative images were taken using a confocal microscopy. Scale bar, 10 μm.