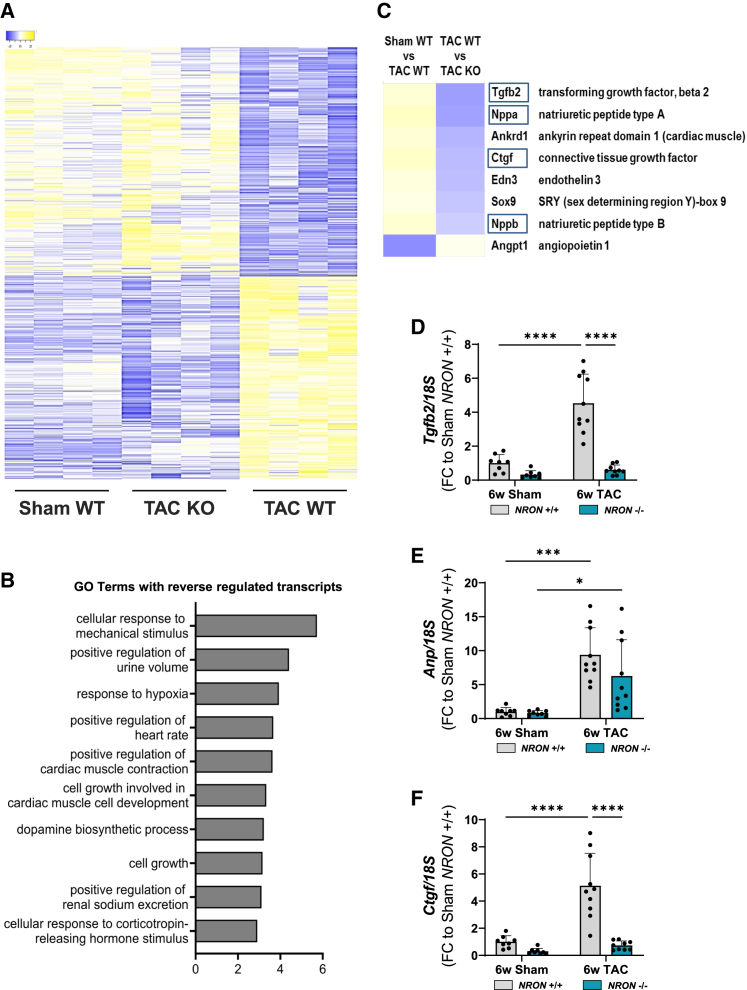
Figure 4.
Transcriptomic changes upon NRON KO
RNA sequencing was performed in hearts from NRON+/+ (WT) and NRON−/− (KO) mice after 6 weeks of Sham or TAC surgery. (A) A heat map shows significantly different mRNAs with reverse regulation between the three groups: Sham WT, TAC WT, and TAC KO. (B) Gene Ontology Biological Process (GOBP) analysis of significantly different mRNAs with reverse regulation between the three groups and a minimum FC of 1.5. n = 4; one-way ANOVA. (C) Cardiac-related genes among the top 100 deregulated mRNAs. (D–F) Levels of Tgfb2 (D), Anp (E), and Ctgf (F) mRNA after 6 weeks of surgery were measured by qPCR and normalized to 18S RNA. All data are mean ± SD; n = 7–12. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. Two-way ANOVA with Tukey's multiple comparisons test.