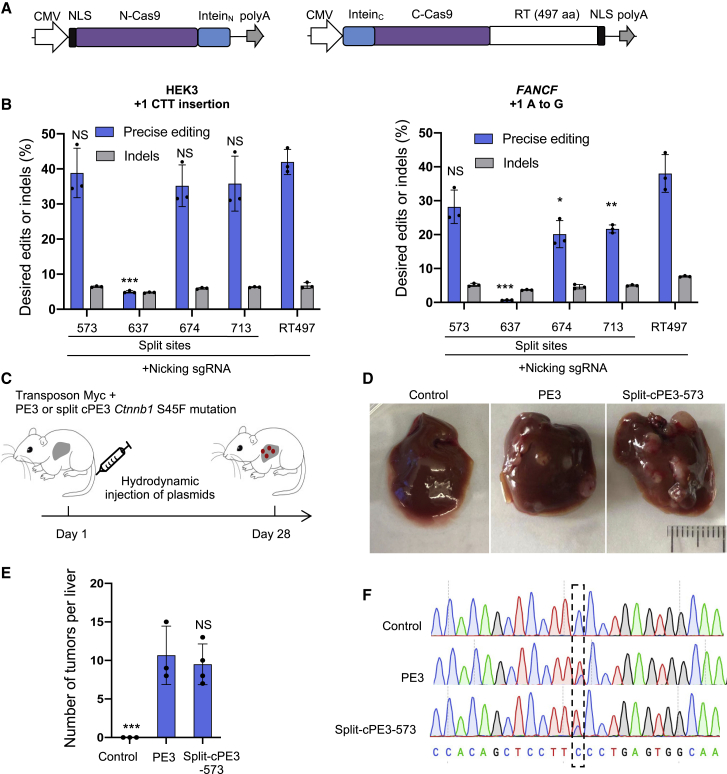
Figure 3.
Development of a split-intein PE3 for in vivo delivery
(A) Map of split-cPE2 plasmids. CMV, cytomegalovirus promoter; NLS, SV40 nuclear localization sequence; N-Cas9 nickase, N-terminal Cas9; C-Cas9 nickase, C-terminal Cas9; poly(A), bGH poly(A). (B) Editing frequencies of split-cPE3 at FANCF and HEK3 in HEK293T cells. Results were obtained from three independent experiments, shown as mean ± SD. (C) PE2 or split-cPE2-573, pegRNA, nicking sgRNA targeting Ctnnb1, MYC transposon, and transposase plasmids were hydrodynamically injected into wild-type FVB mice to generate liver tumors. PE2, non-target pegRNA, MYC transposon, and transposase plasmids were injected in the control group. (D) Representative images of liver tumor burden 28 days after plasmid injection. (E) Mean number of visible tumor nodules. Results were obtained from three (control and PE3) or four (split-cPE3-573) mice, shown as mean ± SD. (F) Sanger sequencing of control liver and representative tumors. NS, not significant; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.