Figure 2. Inactivation of the viral lytic cycle is not sufficient to promote CRISPR immunity.
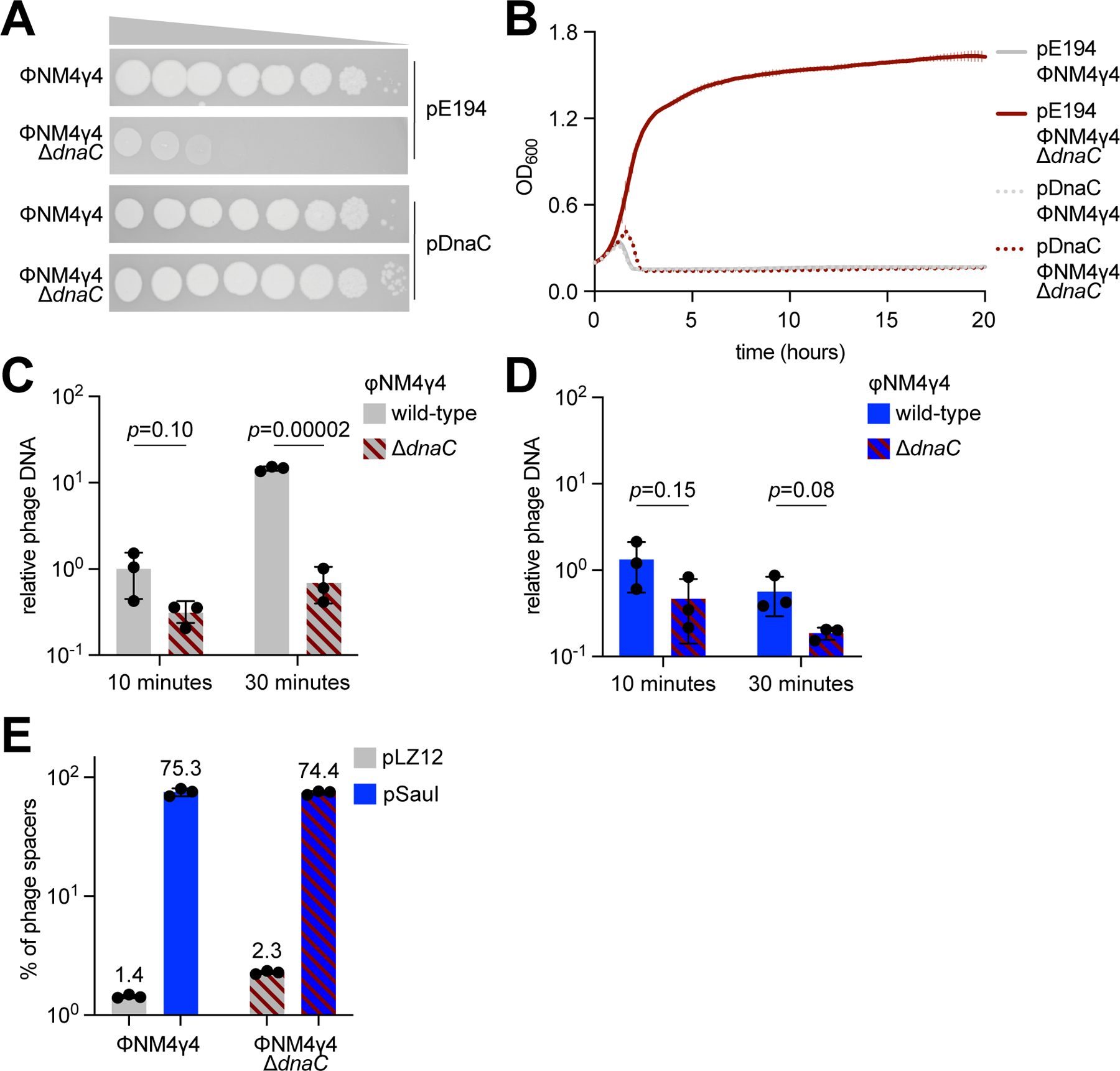
(A) Detection of plaque formation after spotting 10-fold serial dilutions of ΦNM4γ4 or ΦNM4γ4-ΔdnaC phages on top agar plates seeded with S. aureus expressing DnaC or carrying an empty vector control. (B) Growth of staphylococci expressing DnaC or carrying a vector control after infection with ΦNM4γ4 or ΦNM4γ4-ΔdnaC phages, measured as the OD600 of the cultures over time. MOI ~1. Mean of three biological replicates ± SD are reported. (C) Phage DNA quantification via qPCR, relative to host DNA, 10 and 30 minutes post-infection of S. aureus harboring pCRISPR in the absence of restriction with ΦNM4γ4 and ΦNM4γ4-ΔdnaC. MOI ~0.1. Mean of three biological replicates ± SD are reported. (D) Same as (C) but following infection of staphylococci expressing SauI. (E) Quantification of phage-derived spacers, relative to total new spacers, acquired 30 minutes after infection of staphylococci harboring pCRISPR and expressing SauI or carrying a vector control with ΦNM4γ4 or ΦNM4γ4-ΔdnaC phages, via NGS of the CRISPR locus. MOI ~250. Mean of three biological replicates ± SD are reported. See also Figure S2.
