Figure 3. BglII promotes CRISPR spacer acquisition at the restriction site.
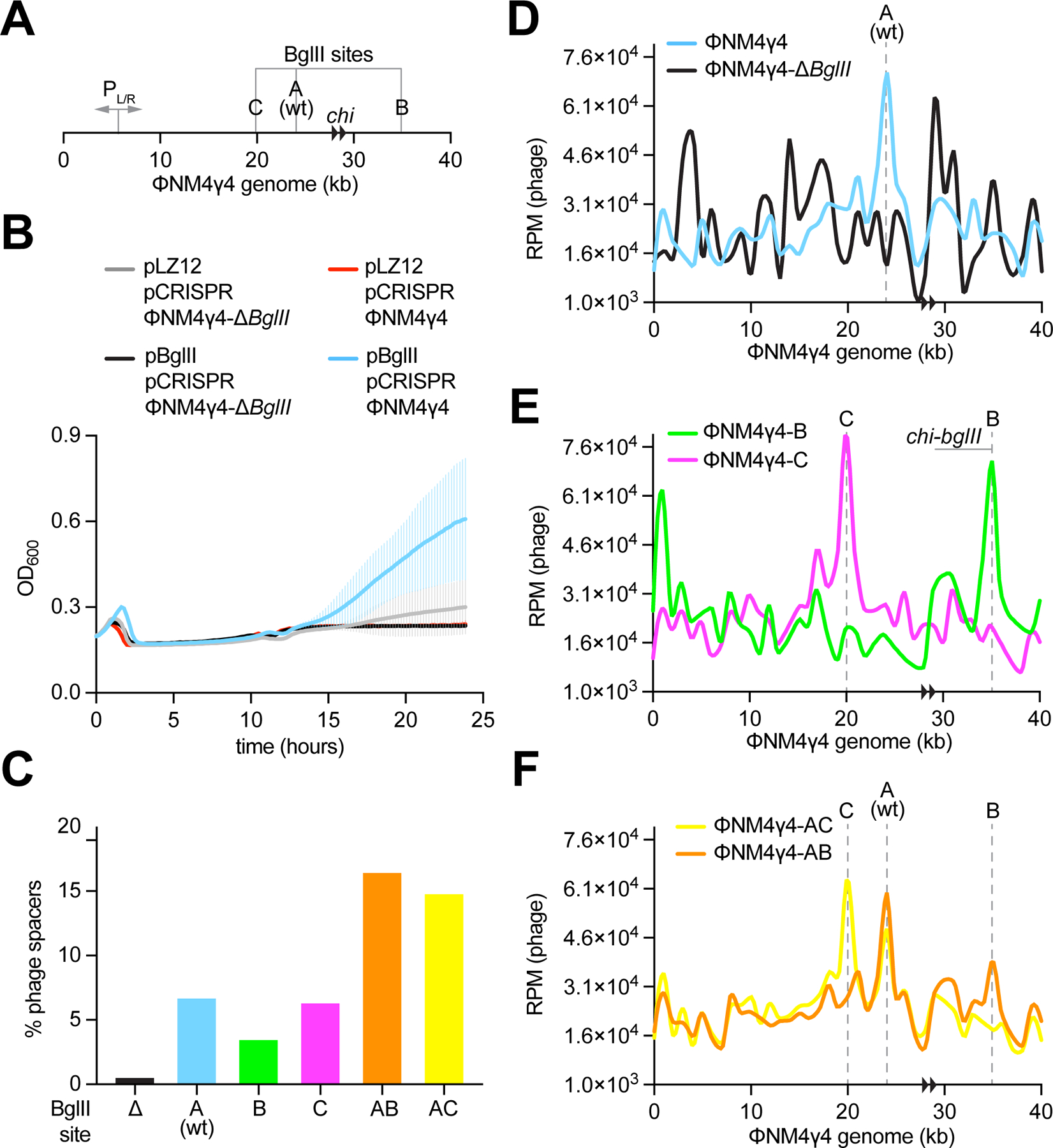
(A) Schematic representation of the ΦNM4γ4 genome showing the BglII sites analyzed in this study, its two chi sites (black arrowheads) and the PL/R bidirectional promoter. (B) Growth of staphylococci harboring pCRISPR and expressing BglII or carrying a vector control after infection with ΦNM4γ4 or ΦNM4γ4-ΔBglII phages, measured as the OD600 of the cultures over time. MOI ~10. Mean of five biological replicates ± SD are reported. (C) Quantification of phage-derived spacers, relative to total new spacers, acquired 30 minutes after infection of staphylococci harboring pCRISPR and expressing BglII or carrying a vector control with ΦNM4γ4 phages containing different BglII sites, via NGS of the CRISPR locus. MOI ~25. (D) Distribution of spacer abundance (measured as RPM of phage-matching reads) obtained in (C) across the ΦNM4γ4 genome, using data from wild-type and ΔBglII infections. (E) Same as (D) using data from ΦNM4γ4-B and ΦNM4γ4-C infections. (F) Same as (D) using data from ΦNM4γ4-AB and ΦNM4γ4-AC infections. See also Figure S3.
