Figure 5. Phage DNA methylation and spacer acquisition occur shortly after phage infection.
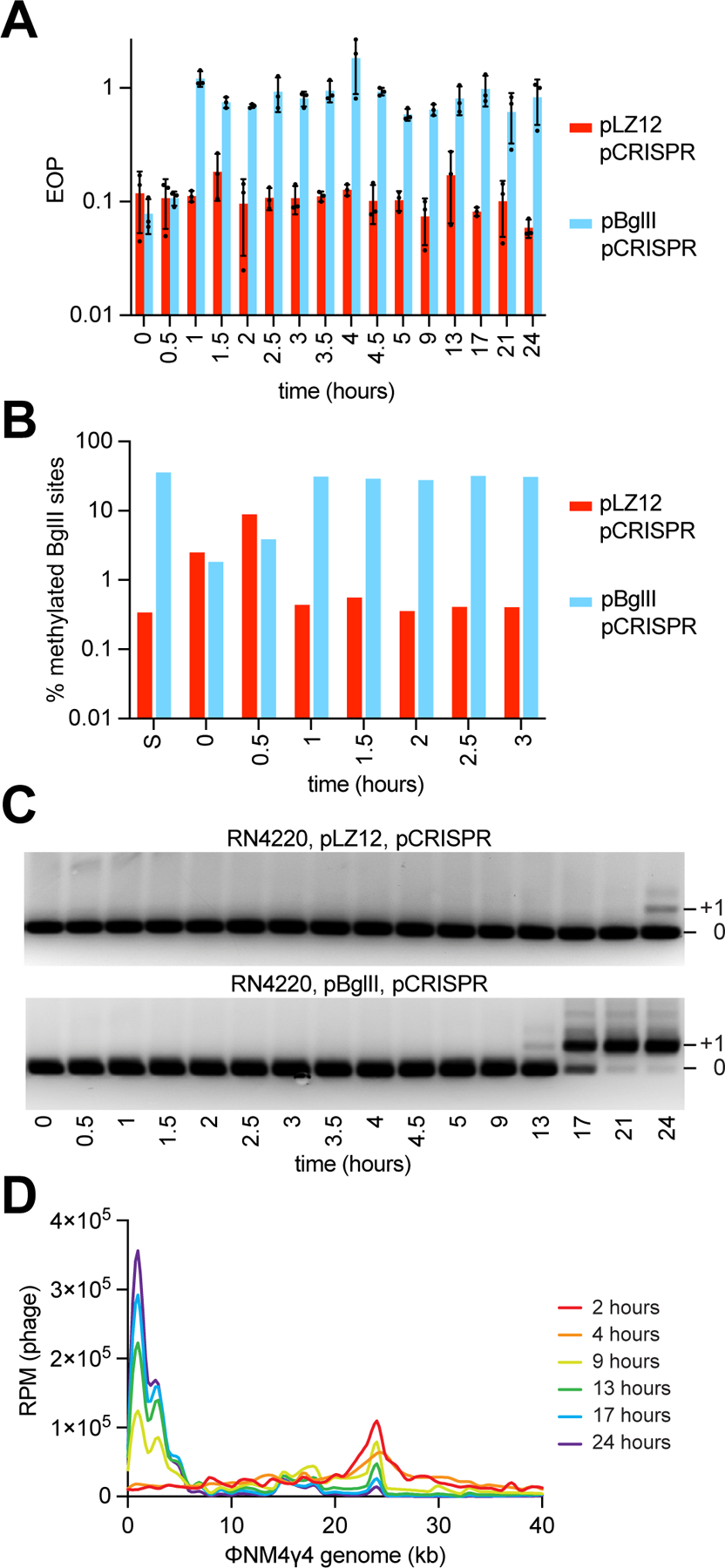
(A) EOP of ΦNM4γ4 phages obtained at different time points following infection (MOI ~1) of S. aureus cells harboring pCRISPR and expressing BglII or carrying a vector control, after plating on lawns of staphylococci expressing BglII, relative to PFUs obtained with cells carrying a vector control. Mean of three technical replicates ± SD are reported. (B) Percent of methylated BglII sites on a unmethylated or methylated ΦNM4γ4 stocks (“S”) and on phages obtained in (A), measured using bisulfite NGS. (C) Agarose gel electrophoresis of PCR products obtained after amplification of the CRISPR array using DNA obtained from the cultures used in (A). (D) Distribution of spacer abundance (measured as RPM of phage-matching reads) obtained after NGS of the CRISPR locus present in the cultures used in (A) across the ΦNM4γ4 genome, using data from the indicated time points. See also Figure S5.
