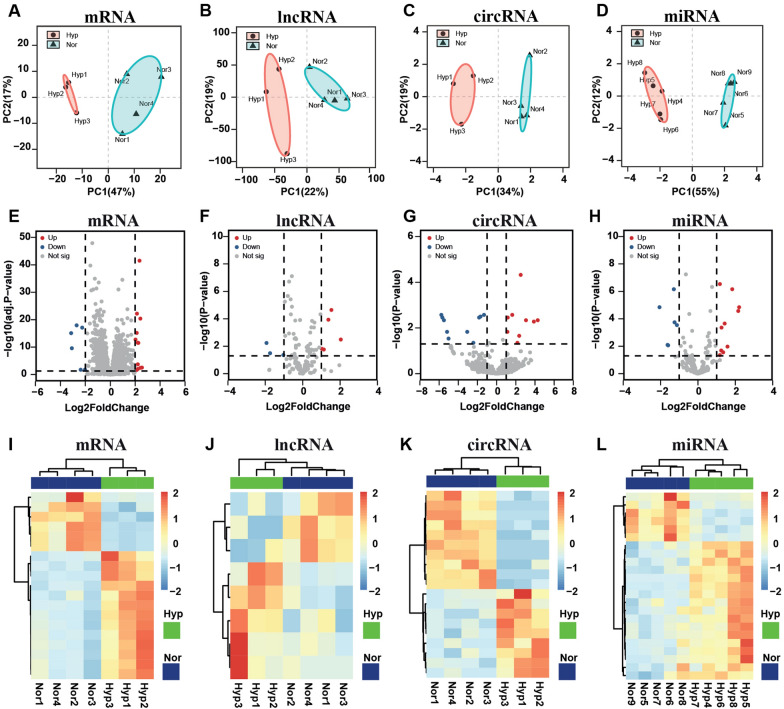
Fig. 3.
Identification of differentially expressed RNAs (DERNAs). Principal component analysis (PCA) of replicates from both hypoxia-induced pulmonary hypertension (HPH) (red) and control (blue) samples. Samples were clustered according to the expression of 500 most variable mRNAs (A), lncRNAs (B), circRNAs (C), and miRNAs (D) in the sequencing dataset. Ellipses represent 95% confidence intervals for the groups. The volcano plot of DEmRNAs (E), DElncRNAs (F), DEcircRNAs (G), and DEmiRNAs (H) between HPH and control samples. Red and blue dots represent downregulated and upregulated DERNAs in HPH rats respectively. The horizontal line represents the value of the padj < 0.05 (E) or p < 0.05 (F–H); the vertical dotted line represents the value of |Log2FoldChange| > 2 (E) or |Log2FoldChange| > 1 (F–H). Expression heatmap of DEmRNAs (I), DElncRNAs (J), DEcircRNAs (K), and DEmiRNAs (L) between HPH and control samples. Unsupervised hierarchical clustering analysis of the DERNAs was performed. Orange color indicates higher expression; blue color indicates lower expression. Nor: normal control rats; Hyp: HPH rats