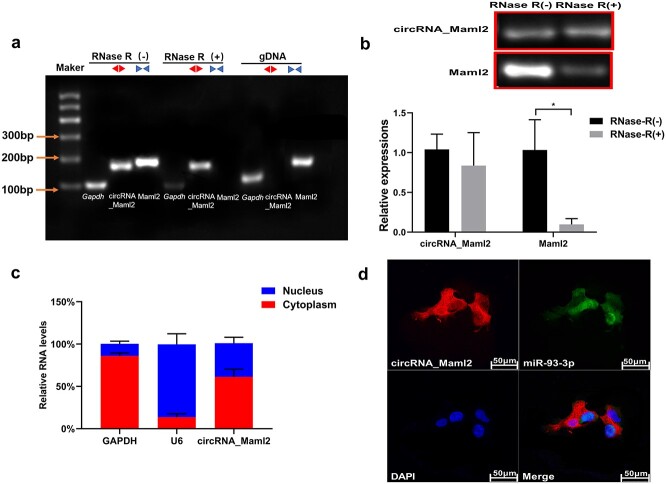
Figure 2.
Identification of circRNA_Maml2 ring structure and location experiment. (a) The existence of circRNA_Maml2 in CT26.wt cells verified by RT-PCR. Different primers can amplify circRNA_Maml2 in cDNA but not genomic DNA (gDNA). Gapdh was used as the linearity control. (b) After RNase R treatment, the relative expressions of circRNA_Maml2 and Maml2 in CT26.wt cells were measured by qRT-PCR assay. (c) The abundance of circRNA_Maml2 in either the cytoplasm or nucleus of CT26.wt cells detected by qRT-PCR. *p < 0.05. (d) FISH was performed to observe the cellular location of circRNA_Maml2 (red) and nucleus (green) in cells (magnification × 200, scale bar= 50 μm). GAPDH Glyceraldehyde-3-phosphate dehydrogenase, qRT-PCR quantitative real-time polymerase chain reaction, FISH fuorescence in situ hybridization