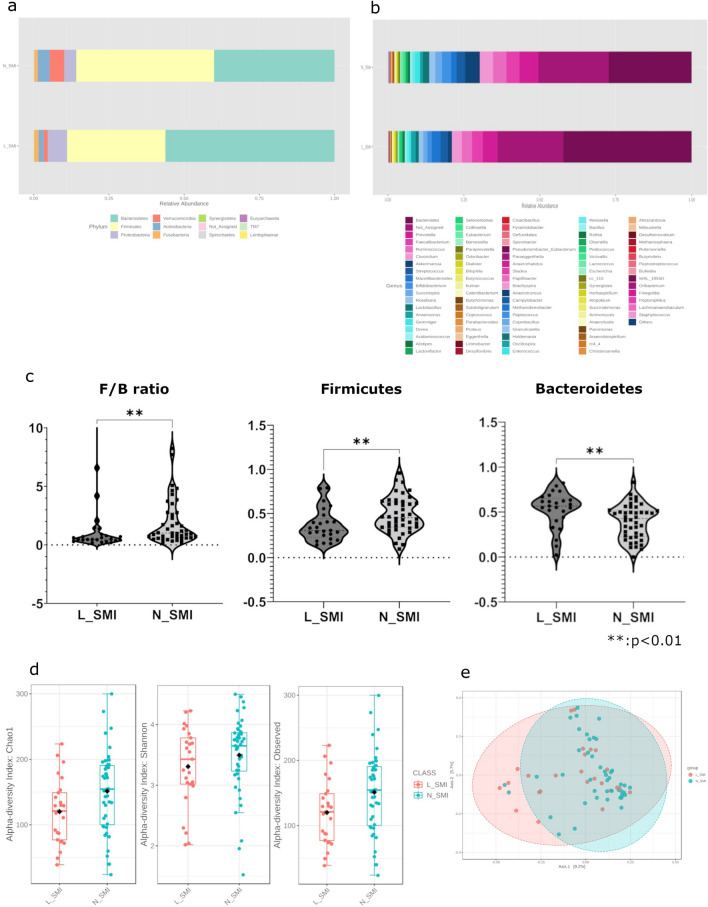
Figure 1.
(a) Proportions of the microbiome at the phylum level. The N-SMI group had a higher proportion of Firmicutes and a lower proportion of Bacteroidetes compared to the L-SMI group. Firmicutes to Bacteroidetes ratio (F/B ratio) was higher in the N-SMI group compared to the L-SMI group. (b) Proportions of the microbiome at the genus level. The phylum Bacteroidetes comprises approximately 90% of the two types of bacteria, Bacteroides and Prevotella. In contrast, the phylum Firmicutes comprises various types of bacteria such as Catenibacterium, Coprobacillus, and Clostridium. (c) F/B ratio and relative abundance of each in the two groups. All two groups were significantly different. F/B ratio (p = 0.0091), Firmicutes (p = 0.0099919, FDR = 0.059951), Bacteroidetes (p = 0.0036818, FDR = 0.044182). One sample in L_SMI contained an outlier of the F/B ratio in the figure. (d) Comparison of alpha diversity. Significantly higher values for N_SMI in Observed species (p = 0.029831) and Chao1 (p = 0.02936); Shannon index (p = 0.2399) was not significantly different. (e) Comparison of β-diversity. No significant difference was found between the two groups. (c.d,e) : Mann–Whitney U test. < Supplementary information for rare names of genus-level bacteria > human: a member of the Rikenellaceae family of bacteria, which includes the gut metagenome as defined by Greengene 13.8. cc_115: Erysipelotrichaceae family, rc4_4: Peptococcaceae family, WAL_1855D: Tissierellaceae family as defined by Greengene 13.8.