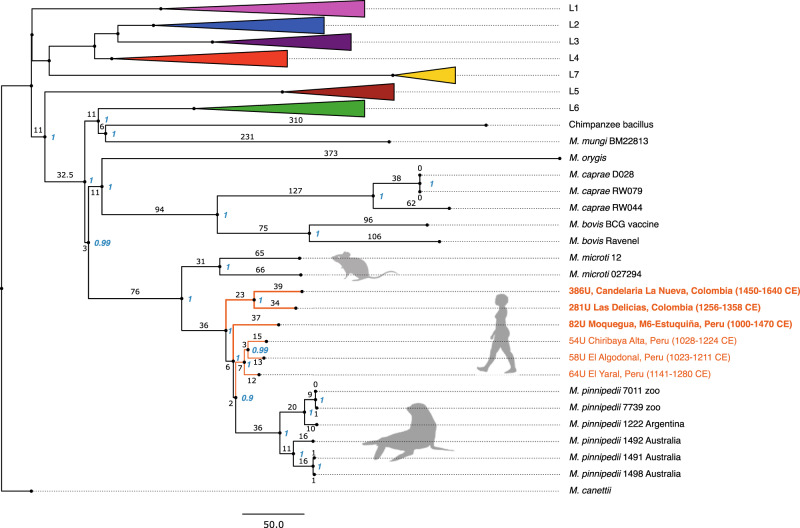
Fig. 2. Maximum Parsimony MTBC phylogeny.
The tree was constructed using the full dataset of 266 MTBC genomes, including the six ancient genomes that are highlighted in orange. Genomes 82U, 281U, and 386U are marked in bold and BAM files filtered for spurious reads were used for SNP calling before tree construction. The tree is based on 14,262 positions out of a possible 44,235, with all missing and ambiguous sites excluded, using 500 bootstrap replicates. Bootstrap support (blue) and branch lengths are marked. Human-adapted lineages 1–5 and 7, and human-associated L6 strains have been collapsed. Our three ancient genomes fall together with other ancient Peruvian genomes within the M. pinnipedii clade.