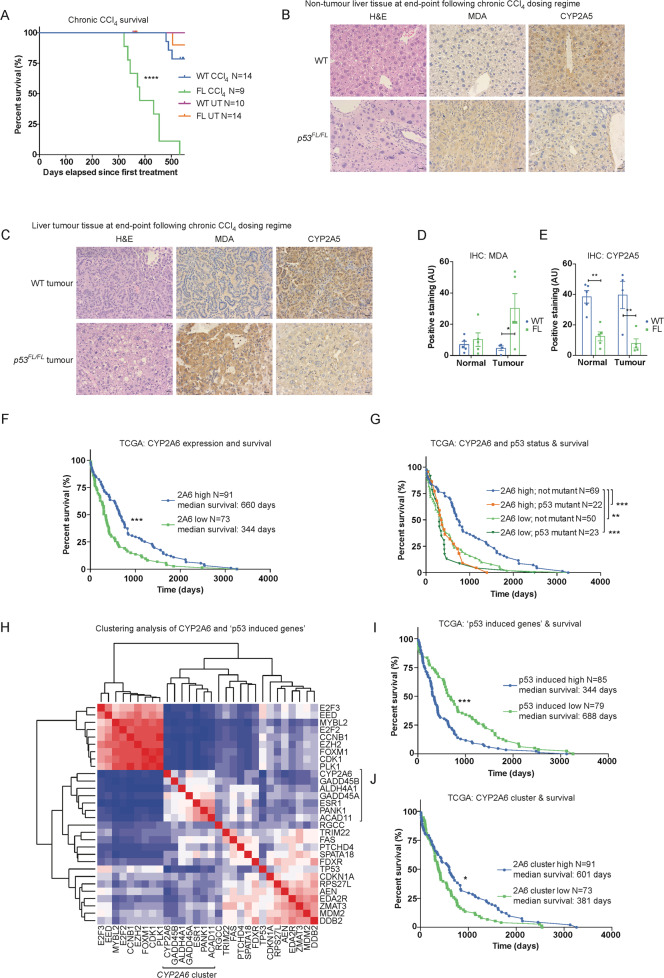
Fig. 4. Liver p53 controls ROS, maintains expression of CYP2A5, and limits tumourigenesis following CCl4-mediated chronic regeneration.
A Survival curve comparing Albumin-Cre; p53WT/WT (WT) and Albumin-Cre; p53FL/FL (FL) mice that were either untreated (UT) or administered the 10-week CCl4 chronic regeneration regime and aged until either reaching clinical endpoint or 550 days after initiation of treatment. Data presented as time (days) since initial CCl4 injection. N = 10 WT and N = 14 FL untreated mice, N = 14 WT and N = 9 FL chronic CCl4-treated mice. Of these, 0/10 WT and 1/14 FL UT mice and 3/14 WT and 9/9 FL CCl4-treated mice reached clinical endpoint within 550 days. All WT tumour mice (3/3) and the majority of FL endpoint tumour mice (5/9) exhibited focal tumour lesions along with substantial non-tumour (normal-like) tissue. These regions were sampled as ‘tumour’ and ‘non-tumour’ tissue for subsequent analyses. Within these cohorts, N = 4 WT and N = 4 FL untreated mice were examined at ~365 days and confirmed to be tumour free. Data analysed using Log-rank (Mantel–Cox) test. ****p < 0.0001. H&E and IHC staining for MDA and CYP2A5 in non-tumour liver tissue (B) and tumour tissue (C) from Albumin-Cre; p53WT/WT (WT) and Albumin-Cre; p53FL/FL (p53FL/FL) mice at clinical endpoint or 550 days post-treatment initiation after prior completion of 10-week CCl4 chronic regeneration regime. Scale bars 20 μm. Images representative of N = 5 mice/group except N = 4 WT tumours (2 from mice at clinical endpoint and 2 small focal tumours identified at experiment endpoint). Additional tumour images included in Supplemental Fig. 3B to illustrate staining from the diversity of tumours observed in the model. Quantification of IHC staining for MDA (D) and CYP2A5 (E) in Albumin-Cre; p53WT/WT (WT) and Albumin-Cre; p53FL/FL (FL) mice from (B/C). Data presented as mean ± SEM and analysed using two-way ANOVA with Holm–Sidak’s multiple comparisons test and multiplicity-adjusted p values. *p < 0.05, **p < 0.01. F Survival curve comparing HCC patients in the TCGA-LIHC dataset with high vs. low expression of CYP2A6 (2A6) on the basis of median survival. N = 91 CYP2A6 high and N = 73 CYP2A6 low patients. Data analysed using the Log-rank (Mantel–Cox) test. ***p < 0.001. G Survival curve comparing HCC patients in the TCGA-LIHC dataset on the basis of TP53 status (loss or mutation of p53 (mutant) vs. WT TP53 (not mutant)) and further breakdown of high vs. low expression of CYP2A6 (2A6) on the basis of median survival. N = 69 2A6 high, not mutant, N = 22 2A6 high, TP53 mutant, N = 50 2A6 low, not mutant, and N = 23 2A6 low, TP53 mutant. Data analysed using the Log-rank (Mantel-Cox) test. **p < 0.01, ***p < 0.001. H Clustering analysis of expression of CYP2A6 related to the previously identified 20 gene ‘p53-Induced Gene Target Expression Signature’ [14] using the TCGA-LIHC dataset. CYP2A6-associated cluster as indicated. For further information, see materials and methods. I Survival curve comparing HCC patients in the TCGA-LIHC dataset on the basis of high vs. low expression of the ‘p53-Induced Gene Target Expression Signature’ [14]. N = 85/79 patients in the high and low groups. This analysis is independent of patient p53 status and includes all data in the TCGA-LIHC dataset. Data analysed using the Log-rank (Mantel–Cox) test. ***p < 0.001. J Survival curve comparing HCC patients in the TCGA-LIHC on the basis of high vs. low expression of the subset of genes within the ‘p53-Induced Gene Target Expression signature’ that clustered with CYP2A6 expression in (H). N = 91/73 patients in the high and low groups. Data analysed using the Log-rank (Mantel–Cox) test. *p < 0.05.