FIGURE 8. Mitofusin-2 knockdown inhibits β2 integrin activation and adhesion of HL60 cells at the same maturation level.
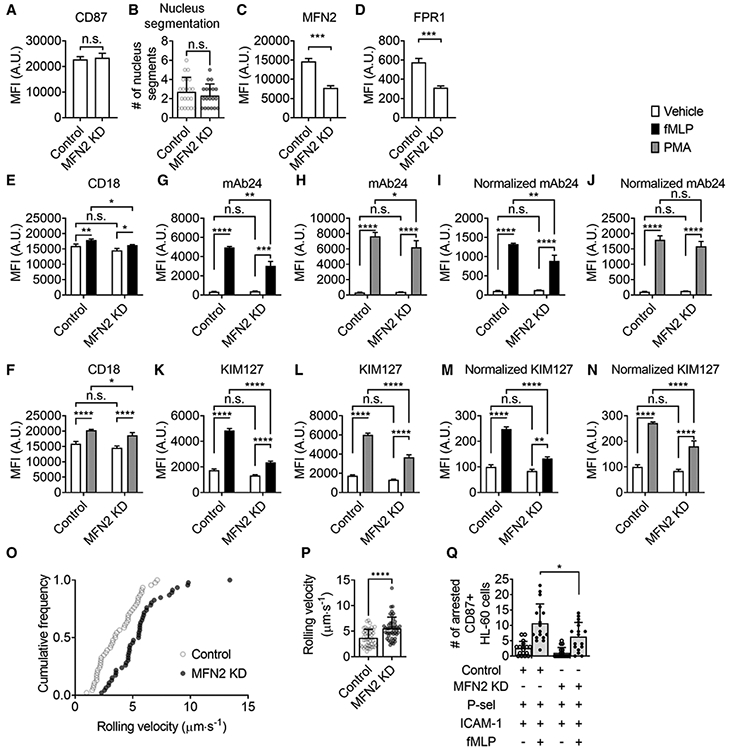
(A) Mean ± SD of surface CD87 expression on CD87high Control and MFN2-knockdown (MFN2 KD) HL60 cells from n = 6 individual experiments. (B) Mean ± SD of nucleus segment number of CD87high Control (n = 21) and MFN2 KD (n = 19) HL60 cells from 9 and 11 independent records, respectively. (C and D) Mean ± SD of MFN2 and FPR1 expression on CD87high Control and MFN2 KD HL60 cells from n = 3 individual experiments. (E and F) Mean ± SD of overall surface CD18 (β2 integrins) expression on CD87high control or MFN2 KD HL60 cells stimulated with FPR1-dependent fMLP (100 nM, at RT for 20 min, closed bars, E), receptor-independent PMA (100 nM, at RT for 20 min, gray bars, F), or vehicle control (open bars) from n = 3 individual experiments. (G-N) Mean±SD of high-affinity (H+, mAb24 staining, G-J) and extended (E+, KIM127 staining, K-N) β2 integrin expression on CD87high control or MFN2 KD HL60 cells stimulated with FPR1-dependent fMLP (100 nM, at RT for 20 min, closed bars, G, I, K, M), receptor-independent PMA (100 nM, at RT for 20 min, gray bars, H, J, L, N), or vehicle control (open bars) from n = 3 individual experiments. In (I and J) and (M and N), the MFI of mAb24 and KIM127 is normalized to overall expression of CD18 (E and F). (O and P) A cumulative histogram (O) and a bar graph (P, mean ± SD, n = 46 cells from 3 individual experiments per group) showing the rolling velocity of CD87high Control or MFN2 KD HL60 cells rolling on the substrate of P-selectin+ICAM-1 under a wall shear stress of 6 dyn/cm2. (Q) The number of arrested CD87high Control or MFN2 KD HL60 cells on the substrate of P-selectin/ICAM-1 with or without the stimulation of fMLP (100 nM) under a wall shear stress of 6 dyn/cm2. Mean ± SD, n = 17 individual records. n.s., non-significant (P > 0.05). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 by Student’s t-test (A-D, P) or 2-way ANOVA followed by Tukey’s multiple comparisons test (E-N, Q). HL60 cells were pre-differentiated with 1.3% DMSO for 7 days
