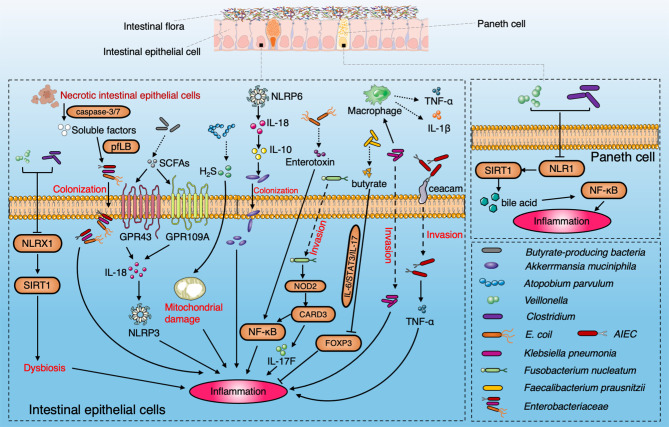
Figure 2.
Diagram summarizing the pathogenic interaction between the gut microbiome and intestinal epithelial barrier in IBD. NLRX1 boosts the dysbiosis to induce the ileac inflammation via SIRT1 signaling pathway with enrichment of Veillonella and Clostridium. SCFAs produced by microbiota induce the activation of NLRP3 through GPR43 and GPR109A and protect colitis through IL-18. A. parvulum produce H2S to induce ileac inflammation by mitochondrial damage. NLRP6 is a key regulator to facilitate the colonization of A. muciniphila via IL-18 and IL-10 to promote IBD onset. AIEC penetrate the mucus layer and adhere to intestinal epithelial cells through FimH and ceacam6. Enterotoxin secreted by E. coli induce IBD by the NF-κB pathway. SIRT1 participates in the inflammation by stimulating Paneth cell to reflect the bile acids metabolism via the NF-κB signaling pathway. Klebsiella pneumoniae invades intestinal epithelial cells and promotes the secretion of IL-1β and TNF by interacting with macrophages. Fusobacterium nucleatum upregulates CARD3 via NOD2 in colonic epithelial cells to activate the IL-17F/NF-κB signaling pathway. Faecalibacterium prausnitzii produce butyrate to inhibit the IL-6/STAT3/IL-17 pathway to activate Foxp3.The soluble factors released by apoptotic intestinal epithelial cells through caspase3/7 facilitate the colonization of Enterobacteriaceae by driving the pyruvate formate-lyase-encoding pflB gene.