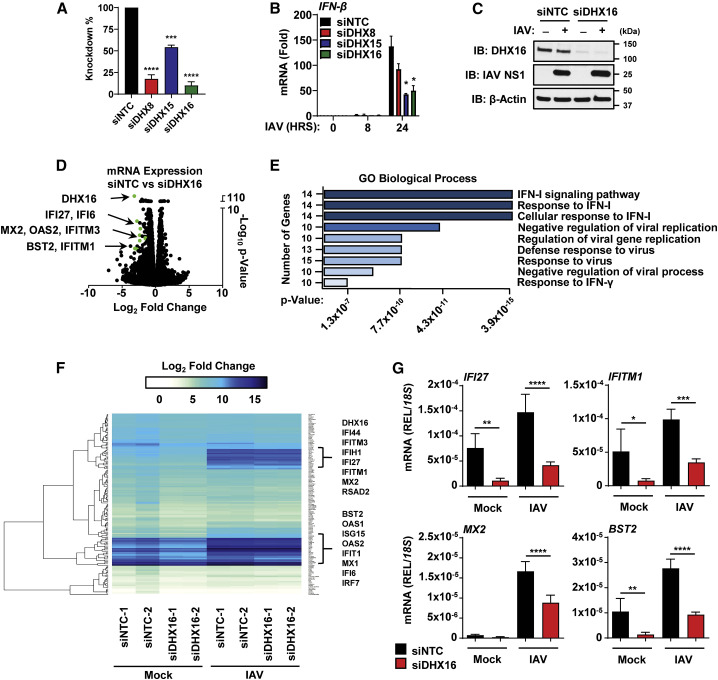
Figure 2.
DHX16 is required for efficient induction of ISGs
(A–C) A549s treated with siNTC or siDHX8/15/16 followed by IAV infection for 24 h (PR8 MOI = 0.1). (A) Silencing efficiency of siRNAs and (B) IFN-β expression (qRT-PCR). (C) Silencing efficiency of DHX16 protein (immunoblot).
(D–F) Next-generation sequencing (NGS) analysis of A549s treated with siRNA and followed by IAV infection as described in (A). (D) Volcano plot of ISGs significantly downregulated in siDHX16 versus siNTC mock samples. (E) Enriched GO terms and pathways (Enrichr) of downregulated transcripts. (F) Heatmap of transcriptomic changes.
(G) Validation of selected ISGs identified in NGS (qRT-PCR).
Data are expressed as means (n = 3) ± SD. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001 (Student's t test or one-way ANOVA with Dunnett's or Sidak's multiple comparisons). Data are representative of 2–3 independent experiments.
See also Figure S1.