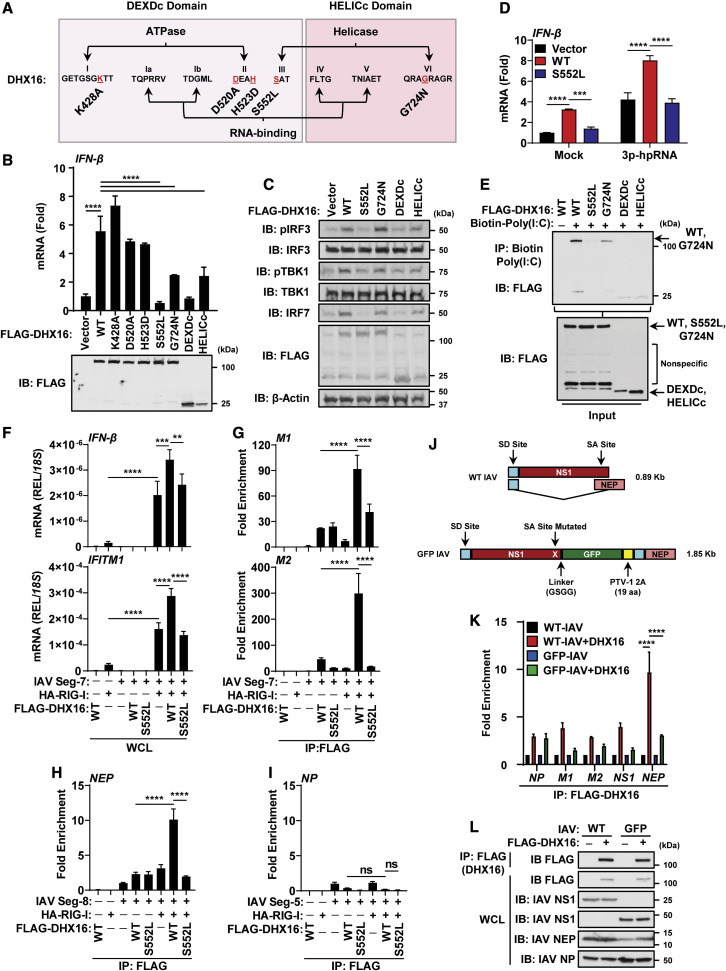
Figure 7.
DHX16 requires an intact motif III for RNA-binding and IFN-I production
(A) Diagram of DHX16 point mutations.
(B) IFN-β expression from A549s transfected with DHX16 (WT or mutants) for 24 h (qRT-PCR and immunoblot).
(C) IFN-I production pathway activation from A549s transfected with DHX16 (WT or mutants) for 24 h (immunoblot).
(D) IFN-β expression from A549s transfected with DHX16 (WT or S552L) and 3p-hpRNA (100 ng/mL) (qRT-PCR).
(E) In vitro RNA-binding ability of DHX16 (WT or mutants) (coIP).
(F–I) HEK293Ts transfected with DHX16 (WT or S552L) and segment 7, 8, or 5 of IAV ± RIG-I for 24 h (RNA-IP). (F) IFN-β and IFITM1 expression from RNA-IP (qRT-PCR). (G–I) Interaction between DHX16 (WT or S552L) and segment 7 (G), 8 (H), or 5 (I) of IAV ± RIG-I (fold enrichment over segment 7, 8, or 5 alone) (RNA-IP).
(J) Diagram of segment 8 from WT and GFP-NS1 IAV PR8 (modified from Manicassamy et al., 2010). WT PR8 expresses NS1/NEP proteins by mRNA splicing (top). GFP-NS1 PR8 expresses both NS1-GFP and NEP proteins from a single mRNA via a ribosome start/stop at the porcine teschovirus-1 2A (PTV-1 2A) site (bottom). Splice donor/acceptor sites (SD/SA).
(K) Interaction between DHX16 and IAV vRNAs from cells infected with WT or GFP-NS1 PR8 for 24 h (PR8 MOI = 0.1) (fold enrichment over IAV alone) (RNA-IP).
(L) Input protein expression from RNA-IP using IAV (WT or GFP-NS1 PR8) (immunoblot).
Data are expressed as means (n = 3) ± SD. ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001 (Student's t test or one-way ANOVA with Tukey's multiple comparisons). Data are representative of 2–3 independent experiments.
See also Figures S6 and S7.