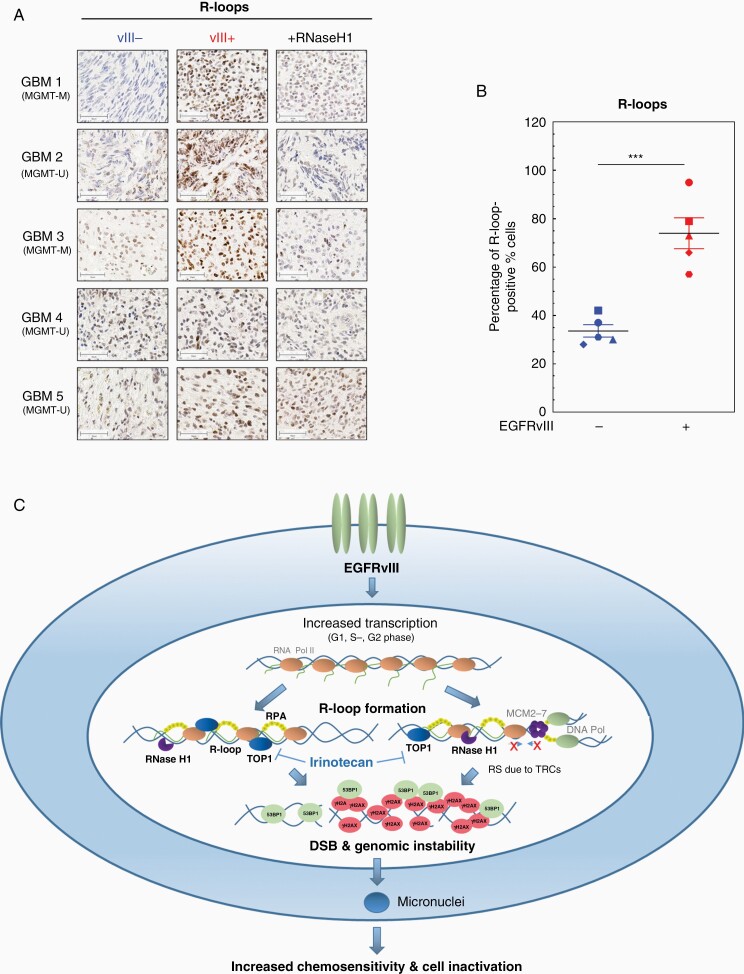
Figure 6.
Increased R-loop accumulation in EGFRvIII-expressing areas in human GBM samples and model for EGFRvIII-induced replication stress. (A and B) Immunohistochemical detection of R-loops in GBM patient samples. (A) Representative pictures for GBM samples 1–5. As a control, samples were incubated with RNase H1 before immunohistochemical staining with S9.6 antibody. EGFRvIII− areas are depicted on the left, EGFRvIII+ areas are displayed in the middle column. EGFRvIII+ areas, which were treated as a control with RNase H1, are shown on the right (scale bars, 50 µm). (B) Quantification of percentage of R-loop-positive cells (n = 5; mean with SEM; P values are obtained by one-tailed Student’s t-test. ***P < .001). (C) Model of how EGRFvIII drives RS: EGFRvIII expression increases transcriptional activity in all cell cycle phases and thereby promotes accumulation of R-loops, which cause DNA damage and genomic instability. R-loop formation during S phase can promote TRCs and can therefore lead to replication fork slowing, stalling, and finally DSB. R-loops in G1 and G2 cells can either be processed by nucleases leading directly to DSB or will form secondary DSB during replication. Both processes result in the activation of the RS response and DDR. If not properly repaired EGFRvIII-induced DSB lead to genomic instability and micronuclei formation and with that cell inactivation.