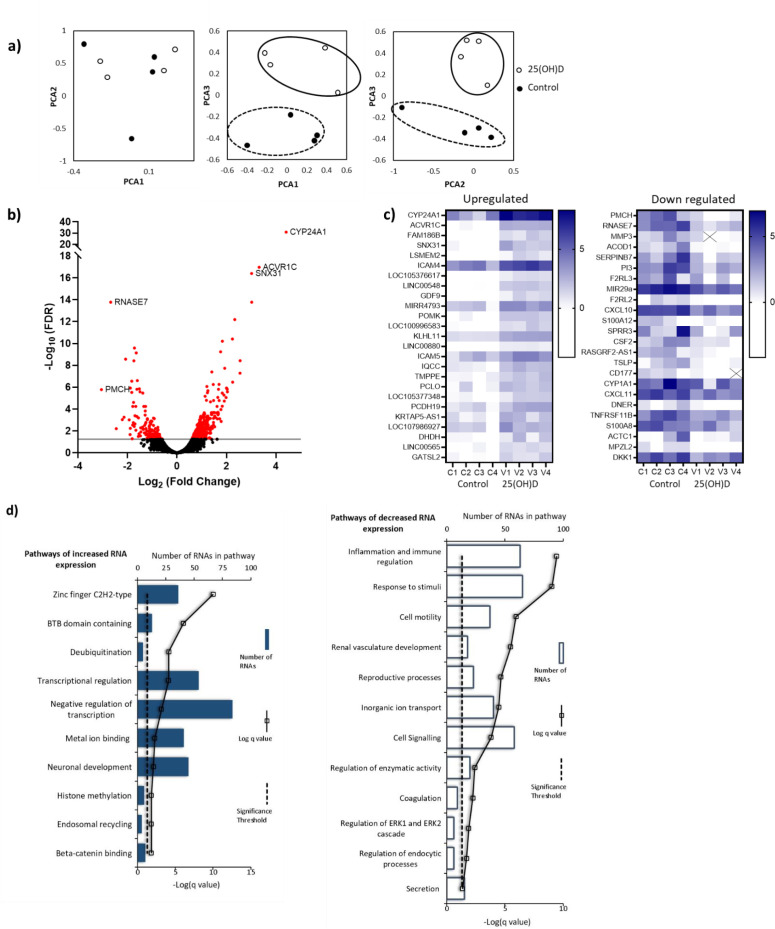
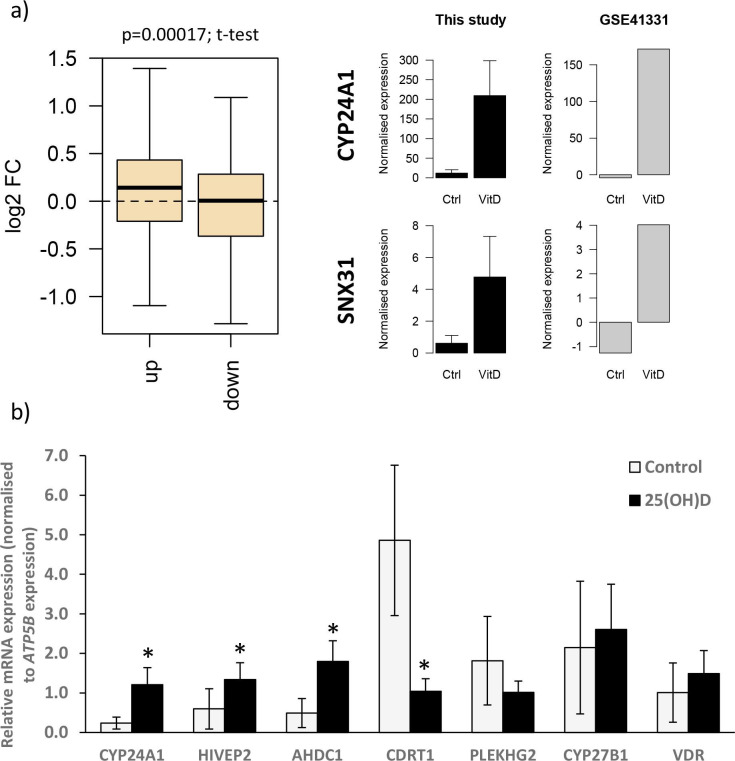
Figure 4. RNA-seq-derived gene signatures of human placental samples following 8 hr 25(OH)D3 incubation.
(a) Principal component analysis indicated clustering of 25(OH)D3-treated samples (clear) compared with control samples (black). (b) Volcano plot showing 493 genes were differentially expressed (red) at a false discovery rate (FDR)-adjusted p-value<0.05 (gray line). (c) Differentially expressed genes with a fold change of 1.5 or above at FDR 0.05 are presented as heatmaps [log2(normalized expression)]. (d) Pathway analysis (ToppGene) of all differentially expressed genes (no fold change cutoff) reveals both up- and downregulation of molecular function and biological process gene pathways following 8 hr 25(OH)D3 incubation. For pathway analysis, significance was adjusted using the Benjamini–Hochberg correction depicted by –log(B&H q-value) with a significance threshold of 1% (dashed line).