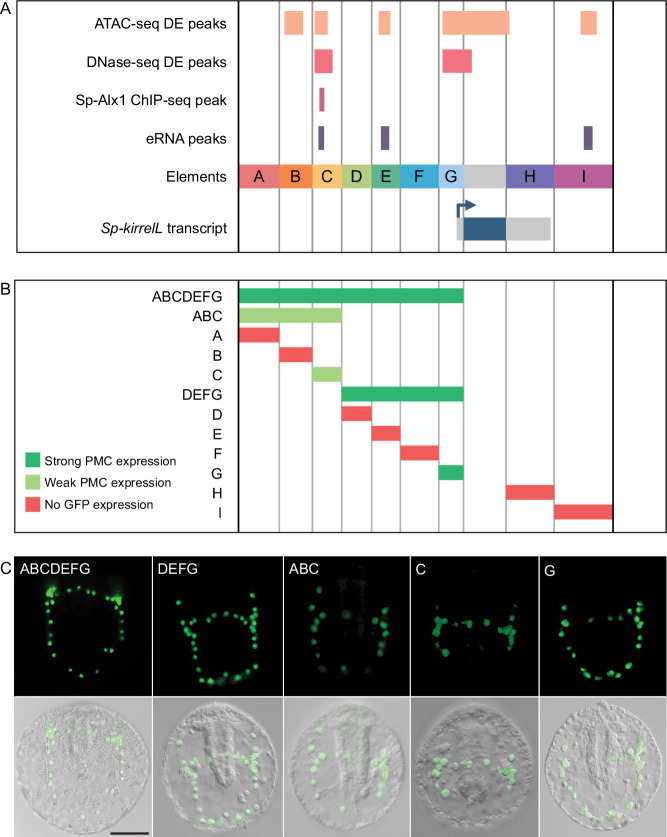
Figure 2. Functional analysis of noncoding genomic sequences flanking Sp-kirrelL to identify cis-regulatory elements (CREs).
(A) Nine putative CREs (labeled elements A–I) were identified based on sequence conservation and previously published datasets (Khor et al., 2021; Khor et al., 2019; Shashikant et al., 2018b). (B) Summary of GFP expression regulated by putative CREs, as assessed by transgenic reporter assays. To be indicated as ‘strong primary mesenchyme cell (PMC) expression’, two criteria were satisfied: (1) more than 1/3 of all GFP-expressing embryos showed expression that was completely restricted to PMCs, and (2) the number of embryos in this class represented >15% of all injected embryos. ‘Weak PMC expression’ was defined similarly except that the number of embryos with expression completely restricted to PMCs represented <15% of all injected embryos. Complete scoring data for all constructs are contained in Figure 2—source data 1. (C) Spatial expression patterns of GFP reporter constructs containing different Sp-kirrelL elements at 48 hr postfertilization (hpf). Top row: GFP fluorescence. Bottom row: GFP fluorescence overlayed onto differential interference contrast (DIC) images. Scale bar: 50 μm.