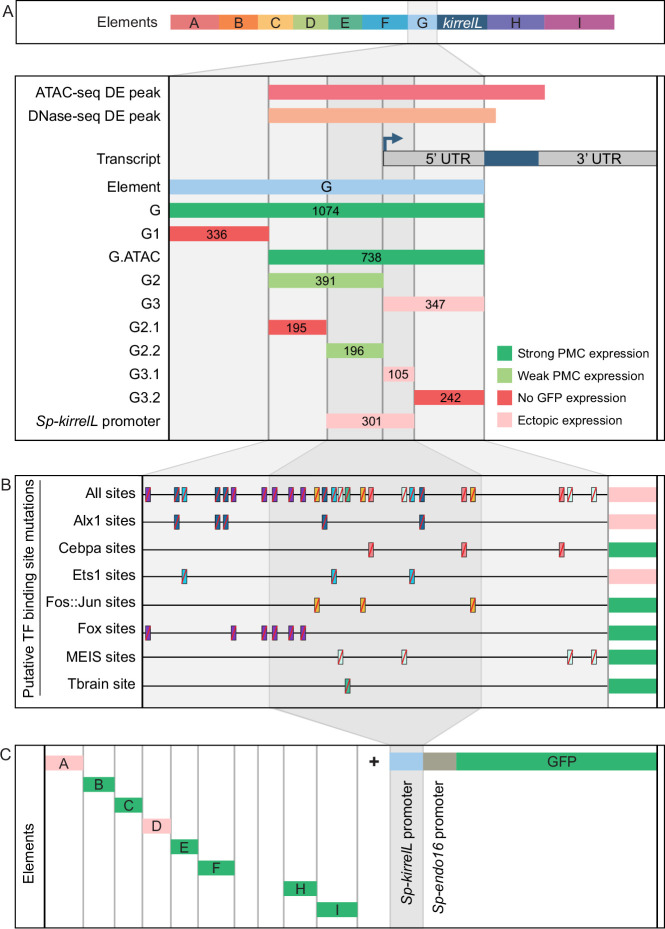
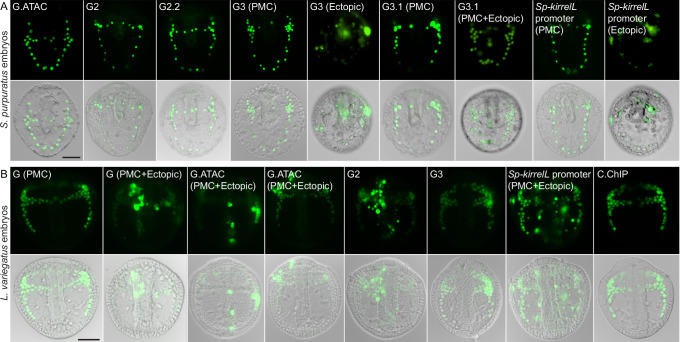
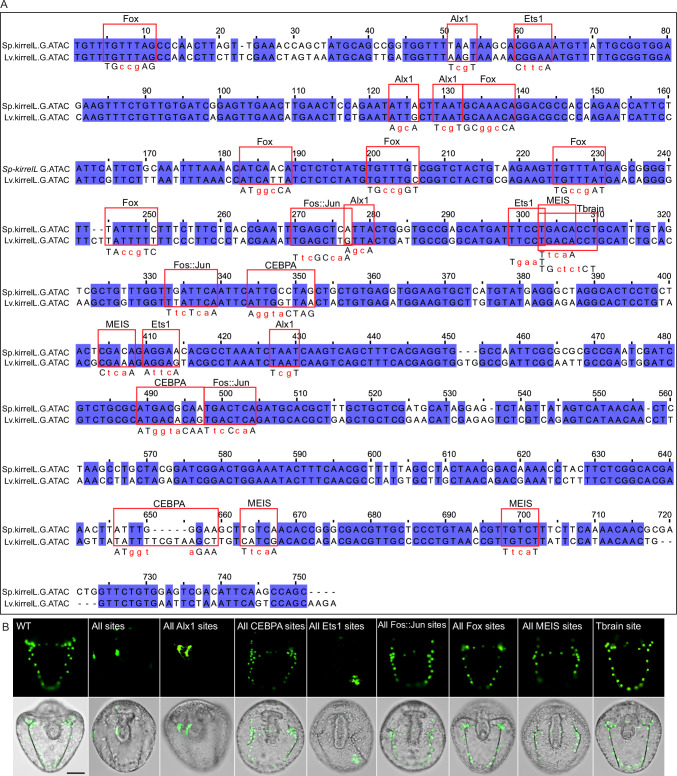
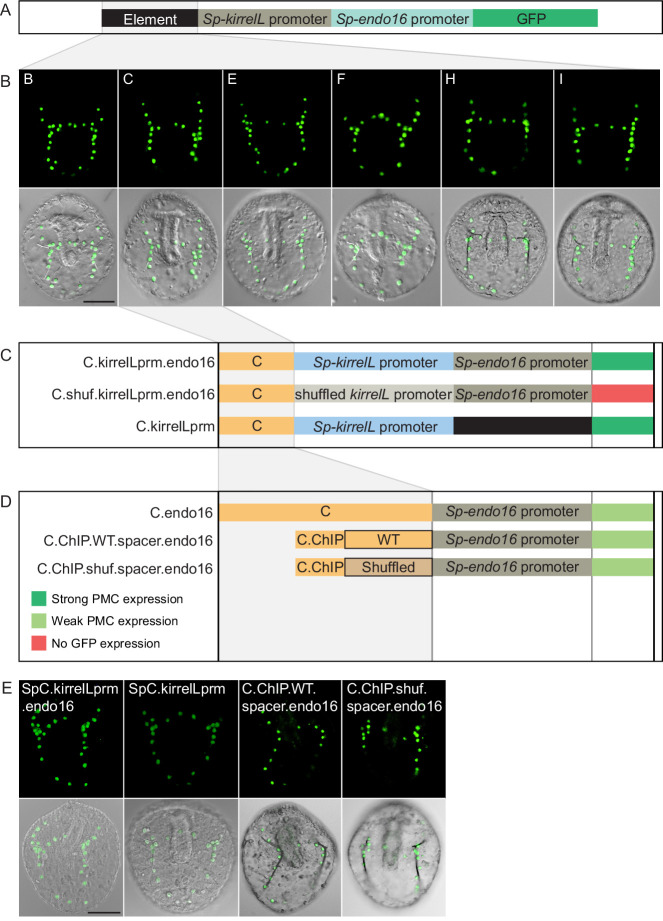
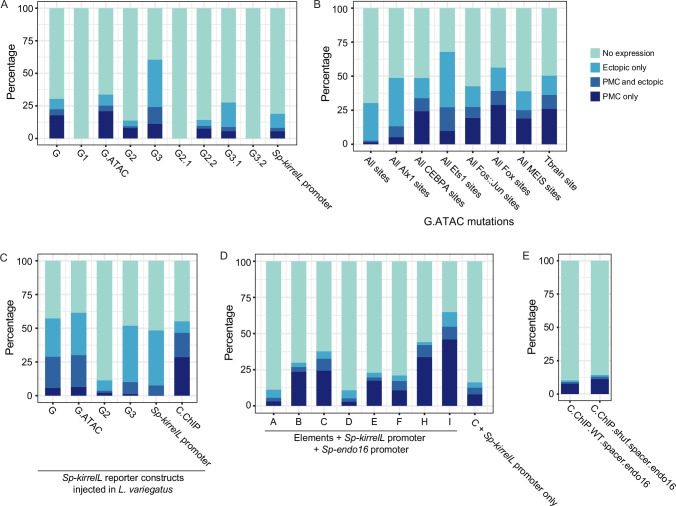
Figure 4. Molecular dissection and mutation of element G.
(A) Summary of GFP expression regulated by element G truncations using EpGFPII reporter constructs. Serial truncation of element G was performed based on boundaries defined by chromatin accessibility and the kirrelL 5′-UTR. Criteria for strong and weak primary mesenchyme cell (PMC) expression are defined in Figure 2. Ectopic expression is defined as majority of injected embryos exhibiting GFP expression in cells other than PMCs. (B) Summary of GFP expression driven by G.ATAC element mutants using EpGFPII reporter constructs. (C) Analysis of element enhancer activity in modified EpGFPII reporter constructs containing the endogenous Sp-kirrelL promoter elements.