ABSTRACT
Increased severity of diseases caused by Coxsackievirus A10 (CV-A10) as well as a large number of mutants and recombinants circulating in the population are a cause of concern for public health. A vaccine with broad-spectrum and homogenous protective capacity is needed to prevent outbreaks of CV-A10. Here, we evaluated cross-neutralization of prototype strain and 17 CV-A10 strains from related manufacturers in mainland China in vitro using 30 samples of plasma collected from naturally infected human adults and 18 sera samples from murine immunized with the above strains of CV-A10. Both human plasma and murine sera exhibited varying degrees of cross-neutralizing activities. Prototype A/Kowalik and sub-genotype C3/S113 were most difficult to neutralize. Among all strains tested, neutralization of S102 and S108 strains by 18 different sera was the most uniform, suggesting their suitability for detection of NtAb titers of different vaccines for avoiding biases introduced by detection strain. Furthermore, among all immune-sera, cross-neutralization of the 18 strains of CV-A10 by anti-S110 and anti-S102 was the most homogenous. Anti-S102 exhibiting higher geometric mean titer (GMT) in vitro was evaluated for its cross-protection capacity in vivo. Remarkably, administration of anti-S102 protected mice from lethal dosage of eight strains of CV-A10. These results provide a framework for formulating strategies for the R&D of vaccines targeting CV-A10 infections.
KEYWORDS: Coxsackievirus A10; cross-neutralization; cross-protection; hand, foot, and mouth disease (HFMD); vaccine candidate
Introduction
Coxsackievirus A10 (CV-A10), a kind of single-stranded RNA (ssRNA) virus with non-enveloped icosahedral stereo-symmetric structure belonging to the enterovirus (EV) genus of the Picornaviridae family of viruses, was first isolated from a patient in the USA in the year 1950.1 Since then, CV-A10 has been shown to be prevalent in Japan,2 Singapore,3 Finland,4 France,5 Taiwan,6,7 and Vietnam,8–10 and has been sporadically isolated in Norway,11 Thailand,12–14 and India.15 Individuals infected with CV-A10 usually suffer from fever, sore throat, hand-foot-mouth disease (HFMD), herpangina, onychomycosis,5,16 and occasionally exhibit severe symptoms like aseptic encephalitis17 and cardiopulmonary failure.18 Interestingly, EV-A strains were considered to be the main causative agents of HFMD in infants and children. Since the introduction of inactivated EV-A71 vaccine in mainland China in the year 2015,19 EV-A71-related cases of lethal HFMD have declined. However, other HFMD pathogens, such as Coxsackievirus A6 (CV-A6) and CV-A10, have become increasingly prevalent worldwide, which has led to a change in the etiological spectrum of HFMD. Infections caused by CV-A10 have exceeded EV-A71 and CV-A16 infections in several countries and regions. CV-A10 has become one of the main causative agents of HFMD as noted in the outbreaks of 20046 and 20077 in Taiwan, 2010 in France,5 and 2012–2013 in Wuhan in mainland China.20 More incidences of severe cases of CV-A10 have been reported after 2009.21–23 Compounding the situation further, recent epidemiological findings have revealed a concerning trend of co-circulation of EV-A71, Coxsackievirus A16 (CV-A16), CV-A6, and CV-A10 strains, which has aroused numerous anxieties in the vaccine industry.
Although vaccination is the most effective method to prevent and control CV-A10 outbreaks, variations in the circulating strains pose a challenge to the success of vaccination programs. An ideal vaccine candidate should possess the ability to elicit a wide spectrum of neutralizing activity, capable of stalling the life cycle of different strains circulating in the population. The titer of the neutralizing antibodies should be sufficiently high to confer protection against different strains. To test the cross-neutralizing ability of the antibodies induced by vaccines being developed and marketed in China, we collected 17 CV-A10 strains from related manufacturers in mainland China. These strains included the main vaccine candidate strains as well as the candidate strains of CV-A10 employed for the detection of the neutralizing antibodies. Using the prototypical strain A/Kowalik as a reference, the susceptibility of the 17 CV-A10 strains to cross-neutralization by 30 samples of plasma collected from adults naturally infected with CV-A10 was evaluated in vitro. We also inoculated murine with the 17 CV-A10 strains and the prototypical A/Kowalik strain individually, collected sera samples and tested them for their ability to cross-neutralize the 18 CV-A10 strains used in this study. Although all samples from humans and murine showed cross-neutralization abilities, they differed in the amount of cross-neutralization titers.
One standard method of testing the effectiveness of vaccines is to estimate the neutralization activity induced by the vaccine. However, the neutralization activity varies with the type of the ”detection strain” used in the neutralization assay. In fact, this is a major issue during the comparison of vaccines and it becomes even more pronounced when vaccines are compared for their ability to provide cross-protection against different strains. In this study, we have attempted to use the MAX/MIN ratio (ratio of maximum titer of neutralizing antibody to the minimum titer) exhibited against each of the CV-A10 “detection strains” by sera collected from murine inoculated with 18 different strains of CV-A10 separately for rationalizing the choice of “detection strains.” Two “detection strains,” S102 and S108, showed a low MAX/MIN ratio, suggesting they were fairly uniformly neutralized by all anti-CV-A10 sera. Furthermore, two CV-A10 strains, S110 and S102, elicited uniform titers of cross-neutralizing antibodies against different CV-A10 variants as judged by the MAX/MIN ratio. Sera from murine inoculated with one of these strains with high geometric mean titer (GMT) and homogenous cross-neutralizing capacities was tested for its ability to confer protection against lethal challenge of different CV-A10 strains in mice. The results highlight the utility of CV-A10 vaccine strains that elicit cross-neutralizing activities of fairly equal and high proportion against different strains in evaluating the effectiveness of the vaccine and the use of titer “detection strains” characterized by low MAX/MIN ratios against different anti-CV-A10 sera for estimating and comparing the titers of neutralizing antibodies elicited by different vaccines.
Materials and methods
Cell
Rhabdomyosarcoma (RD) cells were used for virus culture, virus titer detection, and neutralizing antibody (NtAb) titer detection. MEM solution (Nissui Pharmaceutical Co., LTD.; 5900) containing 10% (v/v) fetal bovine serum (FBS, Gibco; 10099141 C), 2 mM L-glutamine (Gibco; 25030081), and 100 IU/ml penicillin and streptomycin (Cellmax; CPS101.02) was used for cell culture; MEM solution containing 2% (v/v) FBS, 2 mM L-glutamine and 100 IU/ml penicillin and streptomycin was used for virus culture, dilution and NtAb titer detection. Adjust The pH value of above MEM solution to 7.2–7.4 with NaHCO3.
Virus
Prototype A/Kowalik and 17 CV-A10 vaccine candidate strains and detection candidate strains were collected, of which prototype (A) Kowalik, S101 and S105 were maintained in the National Institute for Food and Drug Control (NIFDC). Others were gifts from the Institute of Medical Biology Chinese Academy of Medical Science, the National Vaccine & Serum Institute, the WuHan Institute of Biological Products Co., LTD., Minhai Biotechnology Co., LTD., and Sinovac Biotechnology Co., LTD. Except for Kowalik, 17 CV-A10 vaccine and detection candidate strains were isolated from mainland China from 2014 to 2019. The detailed information of virus titers and 50% lethal dose (LD50) on RD cells and virulence in animals is shown in Table 1. Several strains were 10-fold serially diluted with serum-free MEM to 1 × 10−9, and 1-day-old BALB/c mice were challenged i.p. with 20 μl/mouse (5–8 mice/group). Diluents were set up as negative controls. The health, morbidity, and mortality of suckling mice were observed and recorded daily for 21 d, and the LD50 of these strains was calculated. Five vaccine candidate strains were adapted on Vero cells.
Table 1.
List of CV-A10 strains
| Name | Cell | Time | Place* | Mean of virus titer (lg CCID50/ml) | LD50(lg CCID50/ml) |
|---|---|---|---|---|---|
| Kowalik | RD | 1950 | USA | 7.30 | 4.80 |
| S101 | RD | 2014 | Fujian Province | 8.30 | / |
| S102 | Vero | 2015 | Yunnan Province | 8.48 | 5.83 |
| S103 | RD | 2016 | Jiangsu Province | 8.48 | / |
| S104 | RD | 2016 | Jiangsu Province | 8.60 | 1.72 |
| S105 | RD | 2016 | Hubei Province | 8.78 | 0.95 |
| S106 | RD | 2016 | Hubei Province | 7.04 | / |
| S107 | RD | 2017 | Guangdong Province | 8.00 | 2.67 |
| S108 | Vero | 2017 | Zhejiang Province | 8.30 | 4.18 |
| S109 | RD | 2018 | Beijing City | 8.30 | ∞ |
| S110 | RD | 2018 | Beijing City | 8.48 | 1.33 |
| S111 | RD | 2018 | Beijing City | 8.60 | / |
| S112 | RD | 2018 | Beijing City | 8.30 | / |
| S113 | Vero | 2018 | Hubei Province | 7.13 | / |
| S114 | Vero | 2018 | Jiangsu Province | 7.70 | / |
| S115 | RD | 2019 | Jiangsu Province | 8.60 | 3.00 |
| S116 | RD | 2019 | Hubei Province | 8.30 | 3.64 |
| S117 | Vero | 2019 | Hubei Province | 7.96 | / |
*Kowalik was isolated from USA, others were isolated from mainland China.
CV-A10 sequences with complete VP1 region were downloaded from NCBI. Sequences with clear information such as isolation time and place were sorted out. Annotated VP1 region sequences (894 nt) were extracted for cluster analysis, and the clustering similarity threshold was set at 0.97. In the same isolation year, 1–5 sequences were selected as representative strains from the same sampling country, region, or province, and 297 CV-A10 VP1 gene sequences containing accurate sampling information were obtained. A phylogenetic tree was constructed between the VP1 sequences of 17 CV-A10 vaccine candidate strains and detection candidate strains collected and the curated 297 virus strains. MEGA7 software was used for genotyping. Distance less than 0.15 between groups was considered as the same genotype, and the distance less than 0.08 was considered the same sub-genotype. The representative sequences of each genotype and 17 CV-A10 sequences were selected, and the Maximum Likelihood (ML) method was used to construct the phylogenetic tree. Firstly, the optimal nucleic acid replacement model was calculated as TIM2E+R3 using Iqtree 1.6.2 software. Then, the ML tree was constructed with the bootstrap value of 1000. We tagged the ML tree with Adobe Illustrator CC 2019 23.0 software.
Plasma and sera
Thirty samples of plasma were collected from naturally infected people residing in Heze City, Shandong Province of China, by Taibang Biologic Group. The samples were collected from January to May 2019, preserved and kindly given to us for use in this study. Written informed consent was received from donors’ guardians. The anti-S101, anti-S105, anti-S109, anti-S116, anti-S103, anti-S104, anti-S115, anti-S107, anti-S110, anti-S111, anti-S112, and anti-S108 were sera samples collected from female Wistar rats aged 6 weeks immunized with respective viral strains via intraperitoneal (i.p.) route at a dose of 8.00–8.78 lgCCID50/ml. Immunizations were performed at weeks 0 and 2, and blood samples were collected at week 3, centrifuged at 13,000 g at 4°C for 15 minutes to obtain sera. The sera were stored at −20°C. The anti-S102, anti-Kowalik, anti-S113, anti-S106, anti-S114, and anti-S117 murine sera were donated by the Institute of Medical Biology Chinese Academy of Medical Science, the WuHan Institute of Biological Products Co., LTD., and the Minhai Biotechnology Co., LTD. The anti-S102, anti-Kowalik, anti-S113, and anti-S106 were obtained as mentioned above. The anti-S114 and anti-S117 were obtained from female NIH mice aged 6 weeks using the same protocol.
Cross-neutralization assay
Thirty naturally infected plasma from healthy people and 18 sera samples from murine were used for the cross-neutralization test with 18 CV-A10 strains. The blood samples were diluted at 1:8 with MEM (2% FBS), followed by a 2-fold gradient (1:8 to 1:65536). 50 μl of blood samples with different dilutions were neutralized with 100 CCID50/ml CV-A10 strains at 37°C, 5% CO2 for 2 h, and 100 μl of 1 × 105–1.5 × 105/ml RD cells were added. Cytopathic effect (CPE) was observed after incubation at 35°C, 5% CO2 for 7 days. The neutralization titer was calculated by the Reed-Muench method.
Cross-protection test
BALB/c mice aged 1 day (5–8/group) were challenged by lethal doses (50LD50) of 20 μl via i.p., followed by injection with the diluted protective serum via i.p. within an hour. After 21 days of continuous observation, deaths were recorded to evaluate the passive protective effect of serum in vivo.
Statistical analysis
All statistical analyses were performed with GraphPad Prism 8.02 software. Titers of sera were shown as the largest gradients with no CPE in this article. If the titers of neutralizing antibodies were negative (titer<8), they were set as 1:4 for calculation purposes. Common logarithmic transformation of the NtAb titer raw data was used to calculate the GMT. The value of MAX/MIN referred to the ratio of the max value and the min value of NtAbs of every strain detected sera or serum against strains. The NtAb of each anti-serum against counterpart strain was unified by multiplying a co-efficient and shown as 100 eventually, and NtAbs against other strains were conducted the same calculation. The LD50 was calculated as described by the Reed and Muench method.24
Results
Genotyping
We obtained 17 CV-A10 strains and the prototype A/Kowalik strain from institutions and manufacturers involved in R&D work in China. These strains were isolated from different regions of China between 2014 and 2019 (Table 1). To understand the genetic relationship between the CV-A10 strains and their counterparts, we performed genotype analysis. For this, we first curated CV-A10 sequences containing complete information on the sequence of the VP1 region from NCBI. A total of 297 CV-A10 VP1 gene sequences containing accurate and complete sampling information were obtained. A phylogenetic tree was constructed between the VP1 sequences of 17 CV-A10 strains collected by us and the 297 virus strains curated from NCBI (Figure 1). The results showed that the 17 strains isolated from mainland China during the years 2014–2019 were located in the dominant C3 sub-genotype branch. Furthermore, they showed 91.04–99.66% identity with each other and 75.28–77.18% identity with genotype A. The evolutionary distance between sub-genotype C3 and genotypes A, B, C1, C2, D, E, and F were 0.282, 0.209, 0.147, 0.070, 0.240, 0.186, and 0.206, respectively. Although the evolutionary distance between sub-genotypes C3 and C2 is within 0.08, they were located on different branches.
Figure 1.
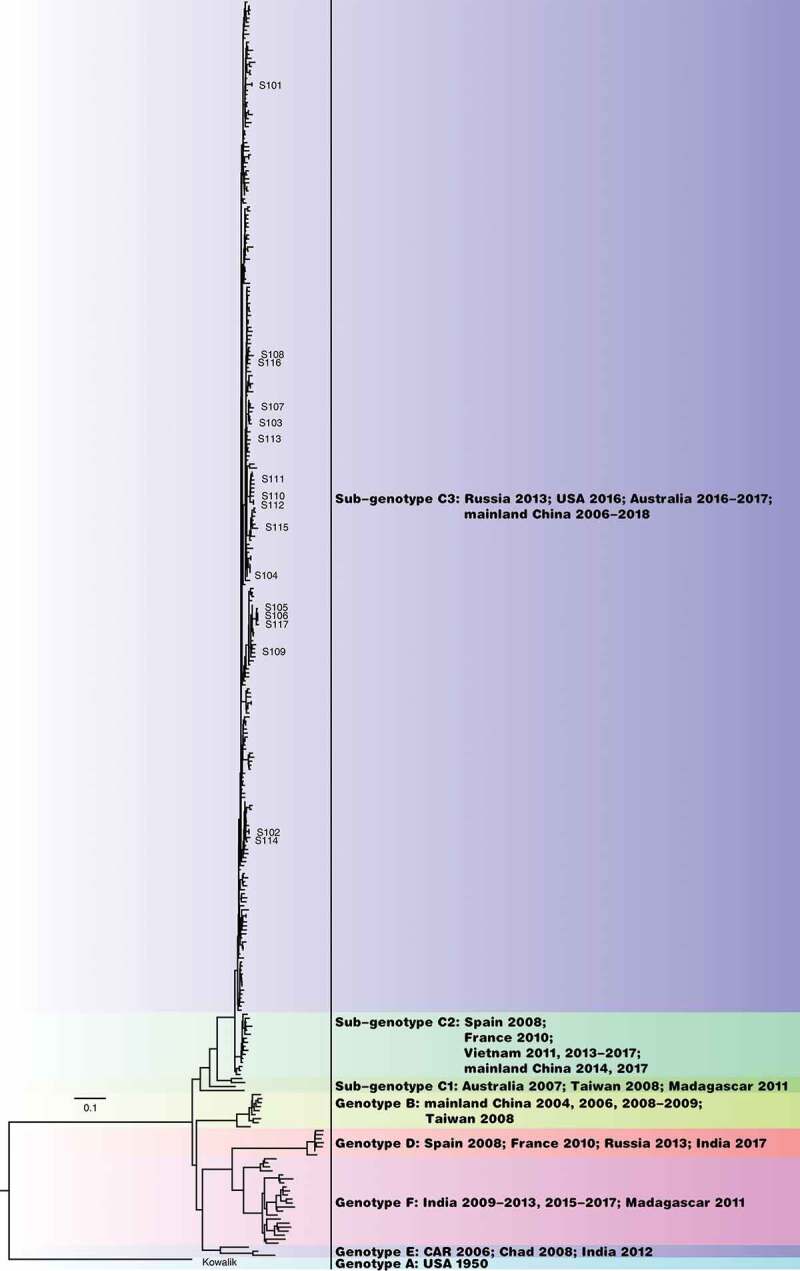
Genotyping of the 17 CV-A10 vaccine and detection candidate strains used in this study.
ML tree showed that the 17 CV-A10 strains are of the sub-genotype C3. The names of strains are marked in the figure.
Cross-neutralization ability of plasma from naturally infected humans
Thirty samples of plasma collected from naturally infected humans living in Heze City of Shandong Province in mainland China from January 2014 to May 2019 were used to test whether they have cross-neutralizing antibodies. Eighteen CV-A10 strains were used for NtAb detection. The common sequence GU947774 in Lu et al.’s study25 and this study indicated that the sub-genotype C3 was prevalent in this region.
The results showed that the neutralization activity of plasma from healthy individuals naturally infected with the virus against the above strains varied, with GMT of neutralizing antibodies against all strains ranging from 14.88 to 124.43. The variation within the C3 sub-genotype ranged from 1.00- to 8.36-fold, and the variation between genotype A and sub-genotype C3 ranged from 1.47- to 4.91-fold. As shown in Figure 2, the order of GMTs against different strains from low to high was as follows: S113 < Kowalik < S109 < S102 < S101 < S114 < S112 < S104 < S110 < S108 < S106 < S117 < S107 < S116 < S103 < S115 < S111 < S105, among which NtAb GMT for S105 was the highest (GMT = 124.43). Furthermore, NtAb GMTs for S113 and Kowalik were the lowest at 14.88 and 25.37, respectively; followed by S109 and S102 (GMT = 37.28, 42.00, respectively), which indicated that the above four strains were the most difficult to be neutralized compared with other endemic strains. Particularly, S113 isolated in Hubei Province of mainland China in 2018 had similar properties with the prototype strain A/Kowalik isolated from the United States in 1950. The largest variation among other 14 strains was 2.96-fold, suggesting a similarity between the strains to some extent.
Figure 2.
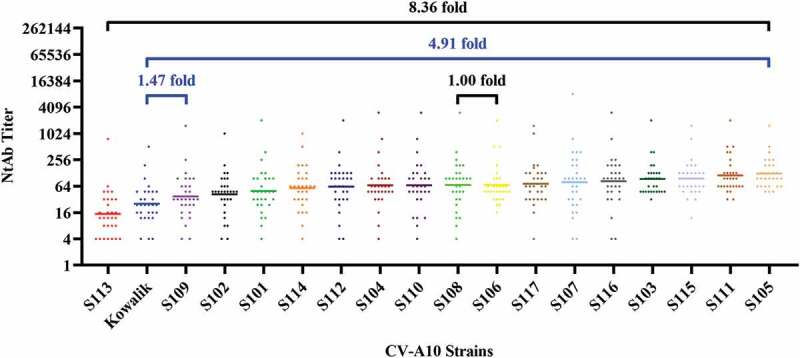
Cross-neutralizing capacity of plasma from humans naturally infected with CV-A10.
The GMTs of 30 human plasma against various strains of CV-A10 ranged from 14.88 to 124.43. This range narrowed down to 1.00–8.36 when GMTs for only strains of sub-genotype C3 were taken into account (shown with black bar). Lastly, the difference between the GMTs for genotype A and sub-genotype C3 strains ranged from 1.47 to 4.91 folds (shown with blue bar).
Cross-neutralization competence of murine sera immunized with CV-A10 strains
Eighteen samples of serum were collected after inoculating murine with 18 CV-A10 strains separately. Each of the anti-CV-A10 sera was tested for its ability to cross-neutralize CV-A10 strains. The results showed that all strains of CV-A10, including the genotype A/Kowalik and the 17 sub-genotype C3 strains, possessed the ability to elicit cross-neutralizing activities, but to a different extent (Figure 3). GMTs of 18 sera detected with 18 strains ranged from 93.52 to 641.35. The variation within sub-genotype C3 was 1.00- to 5.48-fold (GMTS105/GMTS103 = 1.00, GMTS111/GMTS113 = 5.48), and that between genotype A and sub-genotype C3 was 1.25- and 6.86-fold (GMTS113/GMTKowalik = 1.25, GMTS111/GMTKowalik = 6.86). Although the overall trend was similar, there were differences between the GMT ranges of murine sera and human plasma. Prototype A/Kowalik and sub-genotype C3/S113 were analogous as far as the competence to be neutralized by either type of anti-sera. Amongst the C3 strains, S113, which has been recently shown to be prevalent in populations, showed the lowest propensity to be neutralized with GMTs ranging from 2.51 to 8.36 folds lower when compared to other strains of sub-genotype C3.
Figure 3.
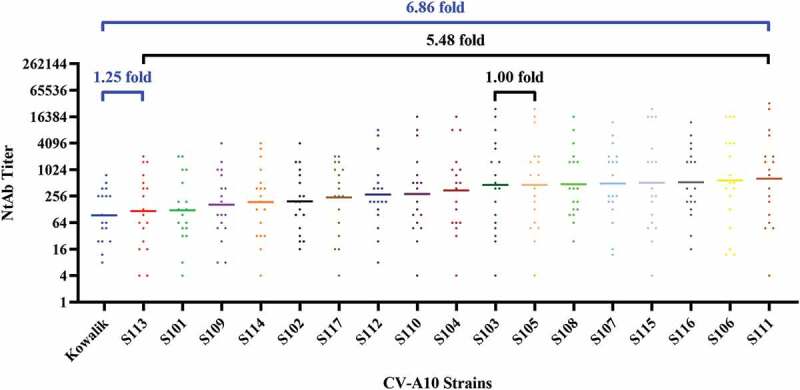
Cross-neutralizing capacity of sera from murine immunized with CV-A10.
The GMTs of 18 murine sera against various strains ranged from 93.52 to 641.35, of which the discrepancy among strains of sub-genotype C3 was 1.00- to 5.48-fold (shown with black bar), and the difference between genotype A and sub-genotype C3 was 1.25- and 6.86-fold (shown with blue bar).
The selection of a uniform viral strain for NtAb detection is important for the comparison and evaluation of immunogenicity of CV-A10 vaccines. In this study, the NtAb titers of each anti-serum against counterpart strain were standardized by converting them proportionately to 100 for comparison while appraising the impacts of each strain for NtAb detection so that the error originating from the different titers of each serum could be evaded. As shown in Table 2, GMTs of the detective strains ranged from 8.50 to 58.33, and the fold was 6.8 over the GMT of Kowalik, which was the lowest, followed by S113, indicating that genotype A was hard to be neutralized compared with sub-genotype C3. Focusing on sub-genotype C3 strains, the GMT of S113 (10.65) was the lowest, and that of S111 (58.33) was the highest, which was 5 times more compared to that of S113. The GMTs observed for the strains starting from the highest are as follows: S111 > S106 > S116 > S115 > S107 > S108 > S105 = S103 > S104 > S110 > S112 > S117 > S102 > S114 > S109 > S101 > S113 > Kowalik.
Table 2.
Susceptibility of CV-A10 strains to Neutralization by Murine Sera in vitro.
| Murine Sera | CV-A10 strains |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Kowalik | S101 | S105 | S102 | S109 | S113 | S106 | S116 | S103 | S104 | S115 | S107 | S114 | S117 | S110 | S111 | S112 | S108 | |
| anti-Kowalik | 100.00 | 75.00 | 800.00 | 75.00 | 75.00 | 37.50 | 800.00 | 75.00 | 600.00 | 200.00 | 600.00 | 75.00 | 200.00 | 150.00 | 200.00 | 800.00 | 200.00 | 150.00 |
| anti-S101 | 25.00 | 100.00 | 75.00 | 100.00 | 150.00 | 50.00 | 50.00 | 400.00 | 150.00 | 100.00 | 75.00 | 600.00 | 50.00 | 50.00 | 75.00 | 150.00 | 75.00 | 200.00 |
| anti-S105 | 0.15 | 0.78 | 100.00 | 0.59 | 0.59 | 0.39 | 100.00 | 1.17 | 100.00 | 6.25 | 100.00 | 1.56 | 2.34 | 12.50 | 3.13 | 150.00 | 1.17 | 1.17 |
| anti-S102 | 25.00 | 50.00 | 37.50 | 100.00 | 100.00 | 50.00 | 75.00 | 300.00 | 50.00 | 100.00 | 25.00 | 200.00 | 100.00 | 50.00 | 75.00 | 75.00 | 37.50 | 150.00 |
| anti-S109 | 66.67 | 16.67 | 50.00 | 66.67 | 100.00 | 200.00 | 133.33 | 533.33 | 50.00 | 33.33 | 50.00 | 400.00 | 33.33 | 66.67 | 50.00 | 50.00 | 100.00 | 100.00 |
| anti-S113 | 16.67 | 2.08 | 3.13 | 33.33 | 50.00 | 100.00 | 0.78 | 100.00 | 2.08 | 3.13 | 3.13 | 100.00 | 2.08 | 1.04 | 3.13 | 4.17 | 12.50 | 133.33 |
| anti-S106 | 16.67 | 50.00 | 200.00 | 6.25 | 2.08 | 33.33 | 100.00 | 25.00 | 400.00 | 133.33 | 400.00 | 16.67 | 50.00 | 33.33 | 133.33 | 533.33 | 50.00 | 33.33 |
| anti-S116 | 25.00 | 3.13 | 3.13 | 100.00 | 100.00 | 100.00 | 3.13 | 100.00 | 1.56 | 4.17 | 3.13 | 400.00 | 2.08 | 2.08 | 4.17 | 3.13 | 12.50 | 266.67 |
| anti-S103 | 6.25 | 2.08 | 400.00 | 6.25 | 2.08 | 1.04 | 33.33 | 4.17 | 100.00 | 50.00 | 100.00 | 3.13 | 8.33 | 66.67 | 12.50 | 266.67 | 12.50 | 6.25 |
| anti-S104 | 6.25 | 25.00 | 9.38 | 50.00 | 50.00 | 25.00 | 50.00 | 150.00 | 37.50 | 100.00 | 37.50 | 150.00 | 49.88 | 25.00 | 75.00 | 18.75 | 100.00 | 100.00 |
| anti-S115 | 0.39 | 4.17 | 100.00 | 3.13 | 1.56 | 1.56 | 66.67 | 16.67 | 100.00 | 66.67 | 100.00 | 8.33 | 12.50 | 6.25 | 33.33 | 133.33 | 12.50 | 6.25 |
| anti-S107 | 25.00 | 0.26 | 0.26 | 16.67 | 16.67 | 25.00 | 0.78 | 100.00 | 0.26 | 0.26 | 0.26 | 100.00 | 0.26 | 0.26 | 0.26 | 0.26 | 0.52 | 100.00 |
| anti-S114 | 16.67 | 266.67 | 400.00 | 25.00 | 25.00 | 4.17 | 1066.67 | 50.00 | 200.00 | 200.00 | 400.00 | 66.67 | 100.00 | 400.00 | 400.00 | 400.00 | 200.00 | 100.00 |
| anti-S117 | 1.17 | 9.38 | 12.50 | 1.56 | 2.34 | 2.34 | 37.50 | 3.13 | 9.38 | 6.25 | 6.25 | 1.56 | 37.50 | 100.00 | 12.50 | 25.00 | 9.38 | 9.38 |
| anti-S110 | 12.50 | 33.33 | 66.67 | 33.33 | 12.50 | 16.67 | 133.33 | 66.67 | 100.00 | 100.00 | 66.67 | 66.67 | 66.67 | 66.67 | 100.00 | 100.00 | 100.00 | 50.00 |
| anti-S111 | 0.29 | 0.59 | 25.00 | 2.34 | 1.17 | 0.05 | 50.00 | 4.69 | 50.00 | 18.75 | 200.00 | 1.56 | 1.56 | 9.38 | 1.17 | 100.00 | 1.56 | 1.17 |
| anti-S112 | 12.50 | 33.33 | 200.00 | 25.00 | 16.67 | 4.17 | 266.67 | 100.00 | 133.33 | 133.33 | 266.67 | 66.67 | 33.33 | 16.67 | 266.67 | 100.00 | 100.00 | 266.67 |
| anti-S108 | 37.50 | 50.00 | 50.00 | 37.50 | 50.00 | 37.50 | 200.00 | 300.00 | 50.00 | 50.00 | 75.00 | 600.00 | 50.00 | 50.00 | 75.00 | 75.00 | 150.00 | 100.00 |
| GMT | 8.50 | 11.17 | 42.18 | 17.68 | 14.91 | 10.65 | 53.30 | 48.43 | 42.18 | 31.59 | 47.35 | 45.26 | 17.06 | 21.84 | 25.98 | 58.33 | 25.33 | 43.84 |
| MAX/MIN | 682.67 | 1024.00 | 3072.00 | 170.67 | 256.00 | 4096.00 | 1365.33 | 455.11 | 2304.00 | 768.00 | 2304.00 | 384.00 | 768.00 | 1536.00 | 1536.00 | 3072.00 | 384.00 | 227.56 |
The values of MAX/MIN of each strain against all sera were calculated to screen out strains homogeneously reactive to any serum. The results demonstrated that MAX/MIN of S113 (4096.00) was the summit, indicating that the relative discrepancies in the neutralization of S113 by different sera were the most evident, whereas those of S102 and S108 were the lowest, which were 170.67 and 227.56, respectively. This meant that the latter two strains showed a minimal relative disparity between neutralizing titers against all sera, and therefore they could be adopted for the detection and appraisal of various vaccines originating from different strains.
Cross-neutralizing capacity of sera from CV-A10-immunized murine
The cross-neutralization potential of anti-CV-A10 sera was studied by using 18 samples of serum collected from murine inoculated with CV-A10 strains and 18 CV-A10 strains as detection strains. As shown in Table 3, most anti-CV-A10 sera collected from murine could neutralize all CV-A10 strains. Amongst the exceptions, while anti-S107 could not neutralize 9 out of the 18 CV-A10 strains tested, anti-S103 and anti-S111 could not neutralize 1 out of 18 strains each. In general, the anti-serum obtained after immunizing murine with genotype A strain showed cross-neutralization capacity against sub-genotype C3 strains, and the anti-sera obtained by immunizing murine with sub-genotype C3 strains could neutralize numerous strains of the same type. Evidently, anti-S107 (GMT = 31.40) and anti-S112 (4019.40) were the poorest and strongest in terms of GMTs of neutralizing antibodies, respectively.
Table 3.
Cross-neutralization ability of Sera from Murine Inoculated with CV-A10 strains
| Murine Sera | CV-A10 strains |
GMT | MAX/MIN | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Kowalik | S101 | S105 | S102 | S109 | S113 | S106 | S116 | S103 | S104 | S115 | S107 | S114 | S117 | S110 | S111 | S112 | S108 | |||
| anti-Kowalik | 256 | 192 | 2048 | 192 | 192 | 96 | 2048 | 192 | 1536 | 512 | 1536 | 192 | 512 | 384 | 512 | 2048 | 512 | 384 | 471.31 | 21.33 |
| anti-S101 | 8 | 32 | 24 | 32 | 48 | 16 | 16 | 128 | 48 | 32 | 24 | 192 | 16 | 16 | 24 | 48 | 24 | 64 | 31.61 | 24.00 |
| anti-S105 | 24 | 128 | 16384 | 96 | 96 | 64 | 16384 | 192 | 16384 | 1024 | 16384 | 256 | 384 | 2048 | 512 | 24576 | 192 | 192 | 715.20 | 1024.00 |
| anti-S102 | 256 | 512 | 384 | 1024 | 1024 | 512 | 768 | 3072 | 512 | 1024 | 256 | 2048 | 1024 | 512 | 768 | 768 | 384 | 1536 | 726.72 | 12.00 |
| anti-S109 | 256 | 64 | 192 | 256 | 384 | 768 | 512 | 2048 | 192 | 128 | 192 | 1536 | 128 | 256 | 192 | 192 | 384 | 384 | 296.91 | 32.00 |
| anti-S113 | 256 | 32 | 48 | 512 | 768 | 1536 | 12 | 1536 | 32 | 48 | 48 | 1536 | 32 | 16 | 48 | 64 | 192 | 2048 | 137.45 | 170.67 |
| anti-S106 | 64 | 192 | 768 | 24 | 8 | 128 | 384 | 96 | 1536 | 512 | 1536 | 64 | 192 | 128 | 512 | 2048 | 192 | 128 | 205.27 | 256.00 |
| anti-S116 | 384 | 48 | 48 | 1536 | 1536 | 1536 | 48 | 1536 | 24 | 64 | 48 | 6144 | 32 | 32 | 64 | 48 | 192 | 4096 | 200.12 | 256.00 |
| anti-S103 | 24 | 8 | 1536 | 24 | 8 | 4 | 128 | 16 | 384 | 192 | 384 | 12 | 32 | 256 | 48 | 1024 | 48 | 24 | 58.91 | 384.00 |
| anti-S104 | 512 | 2048 | 768 | 4096 | 4096 | 2048 | 4096 | 12288 | 3072 | 8192 | 3072 | 12288 | 4086 | 2048 | 6144 | 1536 | 8192 | 8192 | 3523.63 | 24.00 |
| anti-S115 | 96 | 1024 | 24576 | 768 | 384 | 384 | 16384 | 4096 | 24576 | 16384 | 24576 | 2048 | 3072 | 1536 | 8192 | 32768 | 3072 | 1536 | 3305.86 | 341.33 |
| anti-S107 | 384 | 4 | 4 | 256 | 256 | 384 | 12 | 1536 | 4 | 4 | 4 | 1536 | 4 | 4 | 4 | 4 | 8 | 1536 | 31.40 | 384.00 |
| anti-S114 | 64 | 1024 | 1536 | 96 | 96 | 16 | 4096 | 192 | 768 | 768 | 1536 | 256 | 384 | 1536 | 1536 | 1536 | 768 | 384 | 466.88 | 256.00 |
| anti-S117 | 12 | 96 | 128 | 16 | 24 | 24 | 384 | 32 | 96 | 64 | 64 | 16 | 384 | 1024 | 128 | 256 | 96 | 96 | 78.38 | 85.33 |
| anti-S110 | 48 | 128 | 256 | 128 | 48 | 64 | 512 | 256 | 384 | 384 | 256 | 256 | 256 | 256 | 384 | 384 | 384 | 192 | 208.58 | 10.67 |
| anti-S111 | 24 | 48 | 2048 | 192 | 96 | 4 | 4096 | 384 | 4096 | 1536 | 16384 | 128 | 128 | 768 | 96 | 8192 | 128 | 96 | 338.64 | 4096.00 |
| anti-S112 | 768 | 2048 | 12288 | 1536 | 1024 | 256 | 16384 | 6144 | 8192 | 8192 | 16384 | 4096 | 2048 | 1024 | 16384 | 6144 | 6144 | 16384 | 4019.40 | 64.00 |
| anti-S108 | 48 | 64 | 64 | 48 | 64 | 48 | 256 | 384 | 64 | 64 | 96 | 768 | 64 | 64 | 96 | 96 | 192 | 128 | 98.76 | 16.00 |
The value of MAX/MIN of each serum against all strains was calculated to analyze cross-neutralizing homogeneity. As shown in Table 3, MAX/MINs of all strains ranged from 10.67 to 4096.00, indicating that distinct disparity existed among sera. The sequence of MAX/MINs from the lowest to highest is as follows: anti-S110 < anti-S102 < anti-S108 < anti-Kowalik < anti-S104 < anti-S101 < anti-S109 < anti-S112 < anti-S117 < anti-S113 < anti-S114 < anti-S106 < anti-S116 < anti-S115 < anti-S103 < anti-S107 < anti-S105 < anti-S111. The MAX/MINs of anti-S110 and anti-S102 were the lowest with values of 10.67 and 12.00, respectively, which indicated that the cross-neutralization capacity of those two sera was the most homogenous. The MAX/MINs of anti-S105 and anti-S111 were the highest with values of 1024.00 and 4096.00, respectively, suggesting that the cross-neutralization ability of those two sera apparently varied against different strains. To better visualize the cross-neutralization serological data, antigenic cartography was further employed to analyze the serological data. The results of the analysis showed that genotype A strain could be antigenically differentiated from sub-genotype C3 strains (Figure 4).
Figure 4.
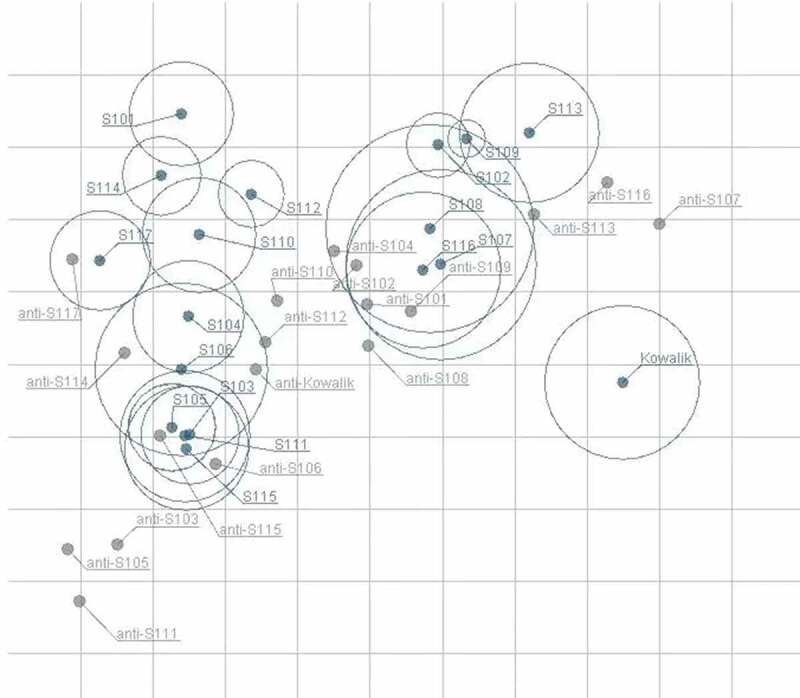
Antigenic map of 18 anti-sera against 18 strains.
The relative positions of viral strains (blue points) and anti-sera (gray points) were adjusted such that the distances between strains and anti-sera in the map represent the corresponding neutralization titers. Outer circles of strains mean standard error. The space between grid lines is 1 unit of antigenic distance, corresponding to a 2-fold dilution of antiserum in the neutralization assay.
Cross-protective experiments in vivo
Anti-S102 possessing better homogeneity (MAX/MIN = 12) and higher GMT than anti-S110 was chosen for demonstration of the protective effect of cross-neutralizing antibodies in warding off CV-A10 infections in vivo. The anti-S102 serum was diluted 16 or 48 folds for validating its cross-neutralizing capacity observed in vitro. One-day-old BALB/c mice were challenged via i.p. with lethal dosages (50LD50) of Kowalik, S105, S116, S115, S107, S110, S108, and S102, separately. Diluted anti-S102 were injected via i.p. route within 1 hour of the challenge. As shown in Figure 5, 16-fold diluted anti-S102 showed a protective effect against lethal doses of 8 CV-A10 strains. The degree of protection conferred by the anti-sera varied from 50% to 100%. The protective effect observed against lethal doses of different strains in ascending order were S107 (50.00%), S115 (50.00%), S110 (66.67%), S105 (66.67%), S116 (71.43%), S108 (83.33%), S102 (83.33%), and Kowalik (100.00%). Thus, anti-S102 showed broad-spectrum cross-protective effect in vivo consistent with the results in vitro.
Figure 5.
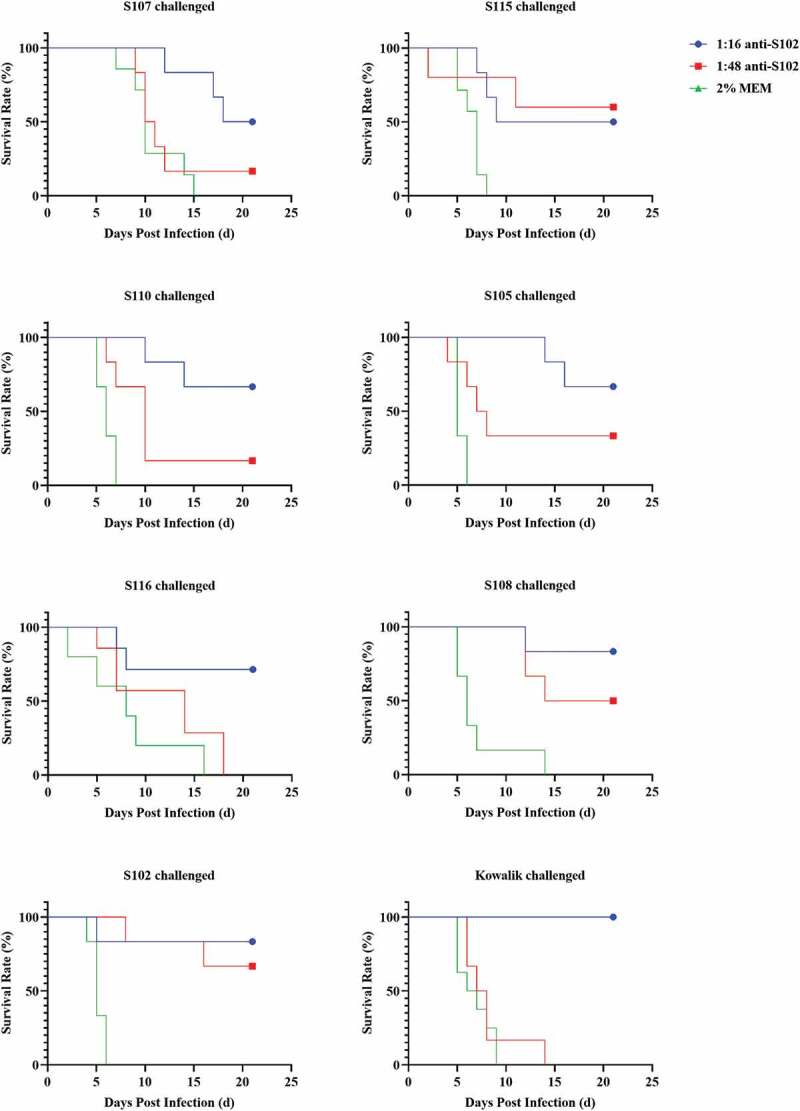
Cross-protective effect of anti – S102 against lethal challenge of 8 CV-A10 strains.
Diluted anti-S102 (16-fold) showed discrepant protective effect against 8 CV-A10 strains, and it ascended from 50% to 100% in the order S107, S115, S110, S105, S116, S108, S102, and Kowalik, with protection against S107 and S116 being both 50% and that against Kowalik being 100%.
Discussion
CV-A10, a neglected pathogen, has been scarcely studied for its pathogenicity. A lack of detailed information on the molecular mechanisms involved in the progression of the disease has hampered vaccine development. Furthermore, the burden of infections and the number of strains have been increasing steadily. All CV-A10 strains are affiliated to one serotype, but they can be divided into several genotypes. However, no uniform standard for this classification has been universally acknowledged. The relatively accepted method is based on the VP1 whole sequence; a difference of more than 15% in the primary sequence of VP1 is regarded as a different genotype, which includes A-D genotypes26,27 with the addition of E-I genotypes later.21,28–30
Kowalik deemed as the prototype (genotype A) is the first isolated strain of CV-A10 in 1950 in America; however, it has not been widely prevalent in populations. In the past couple of decades, CV-A10 has started to attract attention. Molecular epidemiology studies have shown that CV-A10 gene mutation occur routinely in regions of prevalence, which has resulted in different genotypes being prevalent around the world at different times, and even in the same place at the same time. In America and Oceania, the dominant strains during early outbreaks were classified into genotype A and sub-genotype C1, respectively, which eventually converted to sub-genotype C3. Similarly, in Africa, genotype E has been replaced by sub-genotype C1 and genotype F. Sub-genotypes C2, C3, and genotype D were co-prevalent in Europe, even at the same time, while genotypes B-F have been found distributed across Asia. The early dominant genotype found in mainland China was B; but after 2008, sub-genotypes C2 and C3 emerged and dominated.
In this study, the 17 vaccine and detection candidate strains isolated from HFMD cases in seven regions of mainland China like Fujian Province, Yunnan Province, Jiangsu Province, Hubei Province, Guangdong Province, Zhejiang Province, and Beijing City during 2014–2019, were all classified into sub-genotype C3, with the homology in their VP1 sequence ranging from 91.04% to 99.66%. The homology between prototype A/Kowalik and those strains was between 75.28% and 77.18%, suggesting that they are different from genotype A to a large extent.
With such a wide variety of variants circulating in populations and evolving constantly, it is necessary to put countermeasures in place for the prevention and control of outbreaks. Ideally, an effective vaccine should provide protection against all variants. To work through toward this goal, a number of vaccines have been developed in China and several Asia-Pacific countries and regions, including CV-A10 monovalent vaccines,31,32 CV-A6/CV-A10 bivalent vaccines,33 CV-A6/CV-A10/CV-A16 trivalent vaccines,34 and EV-A71/CV-A16/CV-A6/CV-A10 tetravalent vaccines.35 Inactivated and virus-like particle (VLP) vaccines have dominated the vaccine development efforts. Related studies have indicated that monovalent inactivated or VLP CV-A10 vaccines can elicit neutralizing antibodies, with the titers ranging from 287 to 1149.4 against homologous strains, and it slightly reduced against heterologous strains.31,32 For CV-A10 multivalent EV vaccines, no obvious interference manifested among different pathogens,33,35 showing good protective effect in vivo.33,34 These results suggest that inactivated and VLP CV-A10 vaccines and multivalent EV vaccines encompassing CV-A10 have excellent immunogenicity and protective efficacy. However, there are hardly any reports30 on the evaluation of the cross-neutralization capacity of different genotypes of CV-A10 isolated from different locations and during different time points. This information is critical for the selection of strains for formulating the vaccine.
The selection of vaccine strains is a key consideration that has a profound impact on the protective effect of the vaccine. Besides good immunogenicity, wide-neutralizing spectrum, and homogenous reactivity of the elicited NtAbs against other strains are crucial. We studied vaccine candidates of mainland China for these features. The NtAb GMTs of 18 sera samples collected from murine immunized with 17 vaccine strains and the prototype A/Kowalik strain separately were measured and characterized for their cross-neutralization potential. With the exception of three anti-sera, anti-S107, anti-S103, and anti-S111, the rest of the 15 sera could neutralize all CV-A10 strains. In a previous study, Zhou et al.31 showed that NtAb elicited by VLP vaccine originating from genotype A/Kowalik could cross-neutralize CVA10/S0148b and CVA10/S0273b, which were classified into different genotypes, and sera against VLP vaccine could cross-protect against a CVA10/S0148b challenge in vivo. Consistent with those results, cross-neutralization activities with high NtAb titers could be elicited between genotypes by 15 strains in our study. Nonetheless, while anti-S107 could not neutralize 9 out of 18 strains, anti-S103 and anti-S111 could not neutralize 1 out of 18 strains. Interestingly, anti-S107 could robustly neutralize its counterpart strain (titer = 1536), indicating a narrow cross-neutralizing spectrum for anti-S107, which needs further study.
The value of MAX/MIN of each anti-serum against all strains was calculated to compare cross-neutralizing homogeneity. The sequence of MAX/MINs from low values to high were as follows: anti-S110 < anti-S102 < anti-S108 < anti-Kowalik < anti-S104 < anti-S101 < anti-S109 < anti-S112 < anti-S117 < anti-S113 < anti-S114 < anti-S106 < anti-S116 < anti-S115 < anti-S103 < anti- < anti-S105 < anti-S111. Anti-S102 possessing better homogeneity (MAX/MIN = 12) and higher GMT than anti-S110 was subsequently selected for cross-protection studies against eight strains including prototype A/Kowalik in vivo. Diluted anti-S102 (16-fold) showed a protective effect against 8 CV-A10 strains at lethal doses (50LD50), with the protection efficiency ranging from 50.00% to 100.00%, demonstrating that anti-S102 had broad-spectrum and homogeneous cross-protecting capacity in vivo. Previously, EV-A71, another HFMD-related pathogen, was studied for cross-neutralizing capacity using ten sub-genotype C4 strains and one genotype A/Brcr strain. The results revealed that the GMTs of ten sub-genotype C4 strains against all immune sera ranged from 762.44 to 6339.12, and GMT of prototype/Brcr was 1598.37. The differences in GMTs within the strains of sub-genotype C4 ranged from 1.00- to 8.31-fold, and the difference between genotype A and sub-genotype C4 was 1.27- and 3.97-fold.36 Another study on naturally infected healthy adults’ sera showed that the GMTs discrepancy among strains of sub-genotype C4 was 1.02- to 2.06-fold, and the difference between genotype A and sub-genotype C4 was 1.52- and 2.60-fold.37 Similarly, although CV-A10 prototype A/Kowalik prevalence has not been reported as yet, the prototype strain can undergo cross-neutralization as dominant strains regardless of the source of the immune sera, human or murine, indicating that strains from different genotypes can be cross-neutralized. NtAbs of CV-A10 naturally infected healthy adults’ sera can cross-neutralize strains of same or different genotypes, more or less, similar to EV-A71. These results seem to suggest that a number of vaccine strains have the potential to confer cross-protection against different strains of CV-A10.
However, the only anti-sera tested in vivo in our study did not confer complete protection. A plausible reason for this could be that the strains have diverged such that the cross-neutralization capacity of the conserved epitopes has changed. Another reason could be the sub-optimal elicitation of NtAb GMTs in the preliminary murine studies being reported here. A booster dose and or addition of an adjuvant to the vaccine strains during inoculation in murine could perhaps have elicited a more robust and wider cross-neutralization response. Based on previous studies in our laboratory as well as related studies31,32,38,39 showing a good correlation of NtAb titers against CV-A10 with animal protection for the same challenge strain, the results seem consistent with those reported for other enteroviruses such as poliovirus and EV-A71. But for different challenge strains, the correlation of the two was relatively weak. The correlation of cross-protection in vivo with cross-neutralization in vitro showed the same trend between neutralization titer and protection rate for most strains, but some differences existed for strains like Kowalik, S107, and S116. Amongst these, the protective effect against infections by Kowalik was high, but the neutralization capacity was low; whereas the protective effect against S107 and S116 was relatively low, but the neutralization capacity was high. This might be either due to individual variations in animal experiments which made accurate quantitation difficult or due to large variations in virulence of different strains of viruses in animals. The latter reason is more plausible because the animals were challenged uniformly with the virus (50 LD50).
Oral administration of CV-A10 strains in animal models is the most appropriate route of introducing the virus in the animal because of the closer proximity to the route of natural infection. However, one-day-old mice are not easy to work with and feed. To avoid any discrepancies in dosing, the virus was administered via the i.p. route in the current study. Further studies on the impact of the route of administration of the virus are warranted. The prototype strain, Kowalik, shows only 75.28–77.18% identity with sub-genotype C3 strains, indicating differences in the make-up of the genes and possibly the viral proteins that are immunogenic. The results obtained here provide initial guidance on combining CV-A10 strains in the vaccine preparation for combating the strains prevalent in a particular region. However, this idea needs to be tested further.
The results of studies on CV-A10 vaccines produced by manufacturers in China and several Asia-Pacific countries and regions are difficult to compare because of the differences in the use of vaccine candidates, detection strains, and the lack of NtAb standard. Results of our study clearly show that the type of detection strain used in the neutralization assay can have a major impact on the NtAb titer. How do you select a strain for estimating the neutralization titer is a key question in vaccine R&D that has escaped scrutiny. This question becomes even more pronounced with the emphasis shifting on elicitation of cross-neutralizing NtAbs capable of conferring protection against all variants of the pathogen circulating in the population. What kind of strain is best suited to estimate the cross-neutralization potential of the immune response elicited by a vaccine? In an attempt to answer this question and for screening strains that are uniformly susceptible to neutralization, the GMTs of NtAbs of each anti-serum against counterpart strain were converted proportionately to 100 for comparison when appraising the impact of each strain on NtAb detection, so that the error originating from the different titers of each serum could be minimized. Then, NtAb MAX/MINs of all strains against the sera were analyzed with the assumption that a lower value represented better homogeneity of detection.
Theoretically, for this assumption to hold true, the variants need to possess conserved epitopes. This scenario is likely to hold true for closely related variants. A change in conserved epitopes occurs over a period of time, resulting in a possible new genotype or serotype. Assuming that the CV-A10 variants have shared epitopes, it should be possible to find a strain that is relatively uniformly susceptible to anti-sera generated from different CV-A10 strains. Surprisingly, the MAX/MINs ranged from 170.67 to 4096.00, indicating that strains affect NtAb detection. The large difference in the range of MAX/MIN also suggests that there is a difference in the cross-neutralization capacity of the epitopes of the variants. But, a smaller number for MAX/MIN like 170.67 for some strains perhaps suggests that conserved epitopes are more easily detectable in vitro during neutralization assays in such strains than others. Based on the latter assumption, in this study, the NtAb detection discrepancy for S102 and S108 was the lowest. Therefore, these strains can better serve as CV-A10 neutralization antibody detection strains by minimizing the bias introduced by detection strains. Such strains can assist in the evaluation of vaccines and provide a basis for the comparison of different vaccines.
Furthermore, based on the results of our studies, two epidemic strains and Kowalik should be used to evaluate the protection efficacy against a newly emerged variant in vitro and in vivo. The Kowalik with the lowest GMT value of relative NtAb and S111 with the highest GMT value of relative NtAb can be used to evaluate the degree of variation in cross-neutralizing competence in vitro. The S107 with the worst protective effect and Kowalik with the best protective effect can be used to evaluate the degree of variation in cross-protection capacity in vivo. If the GMT of relative NtAb or protective effect against the new variant is lower than the values mentioned above, then the variant should probably be monitored more rigorously.
One of the limitations in this study was the requirement for large amounts of sera for performing the tests. Anti-sera were obtained from two diverse sources – Wistar rats and NIH mice – that were subjected to an identical immunization protocol. Similar accommodations have been made in previous studies.40–42 So, we considered these two sources of sera as similar in properties and that they would have minimal impact on the results. However, additional studies are required to ascertain this.
In summary, the results of our study highlight the potential of vaccine strains to elicit cross-protective neutralizing antibodies. The results provide guidance on the selection of strains for vaccines for broadening the coverage of protection against variants prevalent in a particular region. Our studies also provide an index on selection of detection strains for estimating the NtAb GMTs, which could be a very important consideration during the comparison of vaccines. These results are likely to assist in the development of effective vaccines for combating CV-A10 outbreaks.
Supplementary Material
Acknowledgments
We thank the Institute of Medical Biology Chinese Academy of Medical Science, the National Vaccine & Serum Institute, the WuHan Institute of Biological Products Co., LTD., Minhai Biotechnology Co., LTD., and Sinovac Biotechnology Co., LTD. for CV-A10 strains.
Funding Statement
We thank the National Major Science and Technology Projects of China [2018ZX09737-011].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website at https://doi.org/10.1080/21645515.2021.1978792
References
- 1.Oberste MS, Penaranda S, Maher K, Pallansch MA.. Complete genome sequences of all members of the species Human enterovirus A. J Gen Virol. 2004;85:1597–607. doi: 10.1099/vir.0.79789-0. [DOI] [PubMed] [Google Scholar]
- 2.Itagaki A, Ishihara J, Mochida K, Ito Y, Saito K, Nishino Y, Koike S, Kurimura T. A clustering outbreak of hand, foot, and mouth disease caused by Coxsackie virus A10. Microbiol Immunol. 1983;27(11):929–35. doi: 10.1111/j.1348-0421.1983.tb 00658.x [DOI] [PubMed] [Google Scholar]
- 3.Wu Y, Yeo A, Phoon MC, Tan EL, Poh CL, Quak SH, Chow VTK. The largest outbreak of hand; foot and mouth disease in Singapore in 2008: the role of enterovirus 71 and coxsackievirus A strains. Int J Infect Dis. 2010;14(12):e1076–81. doi: 10.1016/j.ijid.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 4.Blomqvist S, Klemola P, Kaijalainen S, Paananen A, Simonen M-L, Vuorinen T, Roivainen M. Co-circulation of coxsackieviruses A6 and A10 in hand, foot and mouth disease outbreak in Finland. J Clin Virology. 2010;48(1):49–54. doi: 10.1016/j.jcv.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 5.Mirand A, Henquell C, Archimbaud C, Ughetto S, Antona D, Bailly JL, Peigue-Lafeuille H. Outbreak of hand, foot and mouth disease/herpangina associated with coxsackievirus A6 and A10 infections in 2010, France: a large citywide, prospective observational study. Clin Microbiol Infect. 2012. May;18(5):E110–8. doi: 10.1111/j.1469-0691.2012.03789.x [DOI] [PubMed] [Google Scholar]
- 6.Tseng FC, Huang HC, Chi CY, Lin TL, Liu CC, Jian JW, Hsu L-C, Wu H-S, Yang J-Y, Chang Y-W, et al. Epidemiological survey of enterovirus infections occurring in Taiwan between 2000 and 2005: analysis of sentinel physician surveillance data. J Med Virol. 2007;79:1850–60. doi: 10.1002/jmv.21006. [DOI] [PubMed] [Google Scholar]
- 7.Lo SH, Huang YC, Huang CG, Tsao KC, Li WC, Hsieh YC, Chiu C-H, Lin T-Y. Clinical and epidemiologic features of Coxsackievirus A6 infection in children in northern Taiwan between 2004 and 2009. J Microbiol Immunol Infect. 2011;44:252–57. doi: 10.1016/j.jmii.2011.01.031. [DOI] [PubMed] [Google Scholar]
- 8.Hoang MTV, Nguyen TA, Tran TT, Vu TTH, Le NTN, Nguyen THN, Le THN, Nguyen TTH, Nguyen TH, Le NTN, et al. Clinical and aetiological study of hand, foot and mouth disease in southern Vietnam, 2013–2015: inpatients and outpatients. Int J Infect Dis. 2019;80:1–9. doi: 10.1016/j.ijid.2018.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nhan LNT, Khanh TH, Hong NTT, Van HMT, Nhu LNT, Ny NTH, Nguyet LA, Thanh TT, Anh NT, Hang VTT, et al. Clinical, etiological and epidemiological investigations of hand, foot and mouth disease in southern Vietnam during 2015-2018. PLoS Negl Trop Dis. 2020;14:e0008544. doi: 10.1371/journal.pntd.0008544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nhan LNT, Turner HC, Khanh TH, Hung NT, Lien LB, Hong NTT, Nhu LNT, Ny NTH, Nguyet LA, Thanh TT, et al. Economic burden attributed to children presenting to hospitals with hand, foot, and mouth disease in Vietnam. Open Forum Infect Dis. 2019;6. doi: 10.1093/ofid/ofz284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Witso E, Palacios G, Cinek O, Stene LC, Grinde B, Janowitz D, Lipkin WI, Rønningen KS. High prevalence of human enterovirus a infections in natural circulation of human enteroviruses. J Clin Microbiol. 2006;44(11):4095–100. doi: 10.1128/JCM.00653-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chatproedprai S, Theanboonlers A, Korkong S, Thongmee C, Wananukul S, Poovorawan Y. Clinical and molecular characterization of hand-foot-and-mouth disease in Thailand, 2008-2009. Jpn J Infect Dis. 2010 Jul;63(4):229–33 [PubMed] [Google Scholar]
- 13.Linsuwanon P, Puenpa J, Huang SW, Wang YF, Mauleekoonphairoj J, Wang JR, Poovorawan Y. Epidemiology and seroepidemiology of human enterovirus 71 among Thai populations. J Biomed Sci. 2014. Feb 18;21(1):16. doi: 10.1186/1423-0127-21-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Puenpa J, Mauleekoonphairoj J, Linsuwanon P, Suwannakarn K, Chieochansin T, Korkong S, Theamboonlers A, Poovorawan Y. Prevalence and characterization of enterovirus infections among pediatric patients with hand foot mouth disease, herpangina and influenza like illness in Thailand, 2012. PLoS One. 2014. Jun 2;9(6):e98888. doi: 10.1371/journal.pone.0098888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gopalkrishna V, Patil PR, Patil GP, Chitambar SD. Circulation of multiple enterovirus serotypes causing hand, foot and mouth disease in India. J Med Microbiol. 2012;61:420–25. doi: 10.1099/jmm.0.036400-0. [DOI] [PubMed] [Google Scholar]
- 16.Davia JL, Bel PH, Ninet VZ, Bracho MA, Gonzalez-Candelas F, Salazar A, Gobernado M, Bosch IF. Onychomadesis outbreak in Valencia, Spain associated with hand, foot, and mouth disease caused by enteroviruses. Pediatr Dermatol. 2011;28:1–5. doi: 10.1111/j.1525-1470.2010.01161.x. [DOI] [PubMed] [Google Scholar]
- 17.He YQ, Chen L, Xu WB, Yang H, Wang HZ, Zong WP, Xian H-X, Chen H-L, Yao X-J, Hu Z-L, et al. Emergence, circulation, and spatiotemporal phylogenetic analysis of coxsackievirus a6- and coxsackievirus a10-associated hand, foot, and mouth disease infections from 2008 to 2012 in Shenzhen, China. J Clin Microbiol. 2013;51:3560–66. doi: 10.1128/JCM.01231-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He SZ, Chen MY, Xu XR, Yan Q, Niu JJ, Wu WH, Su XS, Ge SX, Zhang SY, Xia NS, et al. Epidemics and aetiology of hand, foot and mouth disease in Xiamen, China, from 2008 to 2015. Epidemiol Infect. 2017;145:1865–74. doi: 10.1017/S0950268817000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mao Q-Y, Wang Y, Bian L, Xu M, Liang Z. EV71 vaccine, a new tool to control outbreaks of hand, foot and mouth disease (HFMD). Expert Rev Vaccines. 2016;15(5):599–606. doi: 10.1586/14760584.2016.1138862. [DOI] [PubMed] [Google Scholar]
- 20.Yang Q, Ding J, Cao J, Huang Q, Hong C, Yang B. Epidemiological and etiological characteristics of hand, foot, and mouth disease in Wuhan, China from 2012 to 2013: outbreaks of coxsackieviruses A10. J Med Virol. 2015;87:954–60. doi: 10.1002/jmv.24151. [DOI] [PubMed] [Google Scholar]
- 21.Tian H, Zhang Y, Shi Y, Li X, Sun Q, Liu L, Zhao D, Xu B. Epidemiological and aetiological characteristics of hand, foot, and mouth disease in Shijiazhuang City, Hebei province, China, 2009-2012. PLoS One. 2017;12:e0176604. doi: 10.1371/journal.pone.0176604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen M, He S, Yan Q, Xu X, Wu W, Ge S, Zhang S, Chen M, Xia N. Severe hand, foot and mouth disease associated with Coxsackievirus A10 infections in Xiamen, China in 2015. J Clin Virol. 2017;93:20–24. doi: 10.1016/j.jcv.2017.05.011. [DOI] [PubMed] [Google Scholar]
- 23.Ji H, Fan H, Lu PX, Zhang XF, Ai J, Shi C, Huo X, Bao C-J, Shan J, Jin Y, et al. Surveillance for severe hand, foot, and mouth disease from 2009 to 2015 in Jiangsu province: epidemiology, etiology, and disease burden. BMC Infect Dis. 2019;19:79. doi: 10.1186/s12879-018-3659-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reed L, Muench H. A simple, method of estimating 50 percent end point. American Journal of Hygiene. 1938; 27, 493–497 [Google Scholar]
- 25.Lu QB, Zhang XA, Wo Y, Xu HM, Li XJ, Wang XJ, Ding S-J, Chen X-D, He C, Liu L-J, et al. Circulation of Coxsackievirus A10 and A6 in hand-foot-mouth disease in China, 2009-2011. PLoS One. 2012;7:e52073. doi: 10.1371/journal.pone.0052073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tian H, Zhang Y, Sun Q, Zhu S, Li X, Pan Z, Xu W, Xu B. Prevalence of multiple enteroviruses associated with hand, foot, and mouth disease in Shijiazhuang City, Hebei province, China: outbreaks of coxsackieviruses a10 and b3. PLoS One. 2014;9(1):e84233. doi: 10.1371/journal.pone.0084233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen AT, Tran TT, Hoang VM, Nghiem NM, Le NN, Le TT, Phan QT, Truong KH, Le NNT, Ho VL, et al. Development and evaluation of a non-ribosomal random PCR and next-generation sequencing based assay for detection and sequencing of hand, foot and mouth disease pathogens. Virol J. 2016;13(1):125. doi: 10.1186/s12985-016-0580-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoa-Tran TN, Nguyen AT, Dao ATH, Kataoka C, Ta HTT, Nguyen HTV, Takemura T, Nguyen TTT, Vu HM, Nguyen TTH, et al. Genetic characterization of VP1 of coxsackieviruses A2, A4, and A10 associated with hand, foot, and mouth disease in Vietnam in 2012-2017: endemic circulation and emergence of new HFMD-causing lineages. Arch Virol. 2020;165:823–34. doi: 10.1007/s00705-020-04536-3. [DOI] [PubMed] [Google Scholar]
- 29.Ji T, Guo Y, Huang W, Shi Y, Xu Y, Tong W, Yao W, Tan Z, Zeng H, Ma J, et al. The emerging sub-genotype C2 of CoxsackievirusA10 associated with hand, foot and mouth disease extensively circulating in mainland of China. Sci Rep. 2018;8(1):13357. doi: 10.1038/s41598-018-31616-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao W, Yue L, Yang T, Li H, Song X, Xie T, He X, Xie Z. A comparative study on biological characteristics of ten coxsackievirus A10 virus strains. Virology. 2021;555:1–9. doi: 10.1016/j.virol.2020.07.003. [DOI] [PubMed] [Google Scholar]
- 31.Zhou Y, Zhang C, Liu Q, Gong S, Geng L, Huang Z. A virus-like particle vaccine protects mice against coxsackievirus A10 lethal infection. Antiviral Res. 2018;152:124–30. doi: 10.1016/j.antiviral.2018.02.016. [DOI] [PubMed] [Google Scholar]
- 32.Shen C, Liu Q, Zhou Y, Ku Z, Wang L, Lan K, Ye X, Huang Z. Inactivated coxsackievirus A10 experimental vaccines protect mice against lethal viral challenge. Vaccine. 2016;34:5005–12. doi: 10.1016/j.vaccine.2016.08.033. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Z, Dong Z, Wang Q, Carr MJ, Li J, Liu T, Li D, Shi W. Characterization of an inactivated whole-virus bivalent vaccine that induces balanced protective immunity against coxsackievirus A6 and A10 in mice. Vaccine. 2018;36(46):7095–104. doi: 10.1016/j.vaccine.2018.09.069. [DOI] [PubMed] [Google Scholar]
- 34.Lim H, In HJ, Lee JA, Sik Yoo J, Lee SW, Chung GT, Choi YK, Chung JK, Cho SJ, Lee J-W, et al. The immunogenicity and protection effect of an inactivated coxsackievirus A6, A10, and A16 vaccine against hand, foot, and mouth disease. Vaccine. 2018;36:3445–52. doi: 10.1016/j.vaccine.2018.05.005. [DOI] [PubMed] [Google Scholar]
- 35.Zhang W, Dai W, Zhang C, Zhou Y, Xiong P, Wang S, Ye X, Liu Q, Zhou D, Huang Z, et al. A virus-like particle-based tetravalent vaccine for hand, foot, and mouth disease elicits broad and balanced protective immunity. Emerg Microbes Infect. 2018;7:94. doi: 10.1038/s41426-018-0094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mao Q, Li N, Yu X, Yao X, Li F, Lu F, Zhuang H, Liang Z, Wang J. Antigenicity, animal protective effect and genetic characteristics of candidate vaccine strains of enterovirus 71. Arch Virol. 2012;157:37–41. doi: 10.1007/s00705-011-1136-3. [DOI] [PubMed] [Google Scholar]
- 37.Gz MAOQY, Cs HAO, YU D, Gao F, Ch DONG, WQ AN, Gao Q, ZP XIE, Xl LI, ZL LIANG. Screening and preparation of candidate standard strain for determination of neutralizing antibody against enterovirus 71. Chin J Biologicals. 2012;25:725–29. [Google Scholar]
- 38.Sun S, Gao F, Mao Q, Shao J, Jiang L, Liu D, Wang Y, Yao X, Wu X, Sun B, et al. Immunogenicity and protective efficacy of an EV71 virus-like particle vaccine against lethal challenge in newborn mice. Hum Vaccin Immunother. 2015;11(10):2406–13. doi: 10.1080/21645515.2015.1053675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu CK, Chen CC, Chen CL, Wang JR, Liu CC, Yan JJ, Su IJ. Neutralizing antibody provided protection against enterovirus type 71 lethal challenge in neonatal mice. J Biomed Sci. 2000. Nov-Dec;7(6):523–8. doi: 10.1007/BF02253368 [DOI] [PubMed] [Google Scholar]
- 40.Mizuta K, Aokia Y, Sutoa A, Ootani K, Katsushima N, Itagaki T, Ohmi A, Okamoto M, Nishimura H, Matsuzaki Y, et al. Cross-antigenicity among EV71 Ss from different genogroups isolated in Yamagata, Japan, between 1990 and 2007[J]. Vaccine. 2009;27:3153–58. doi: 10.1016/j.vaccine.2009.03.060. [DOI] [PubMed] [Google Scholar]
- 41.Zhang H, An D, Liu W, Mao Q, Jin J, Xu L, Sun S, Jiang L, Li X, Shao J, et al. Analysis of cross-reactive neutralizing antibodies in human HFMD serum with an EV71 pseudovirus-based assay. PLoS One. 2014. 25;9(6):e100545. doi: 10.1371/journal.pone.0100545. eCollection 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu P, Yuan YD, Cui BP, Huo Y, Bian L, Chen L, Liu S, Wang C, Xu Y, Tedcastle A, et al. Cross-antigenicity between EV71 sub-genotypes: implications for vaccine efficacy. Viruses. 2021;13(5):720. doi: 10.3390/v13050720. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


