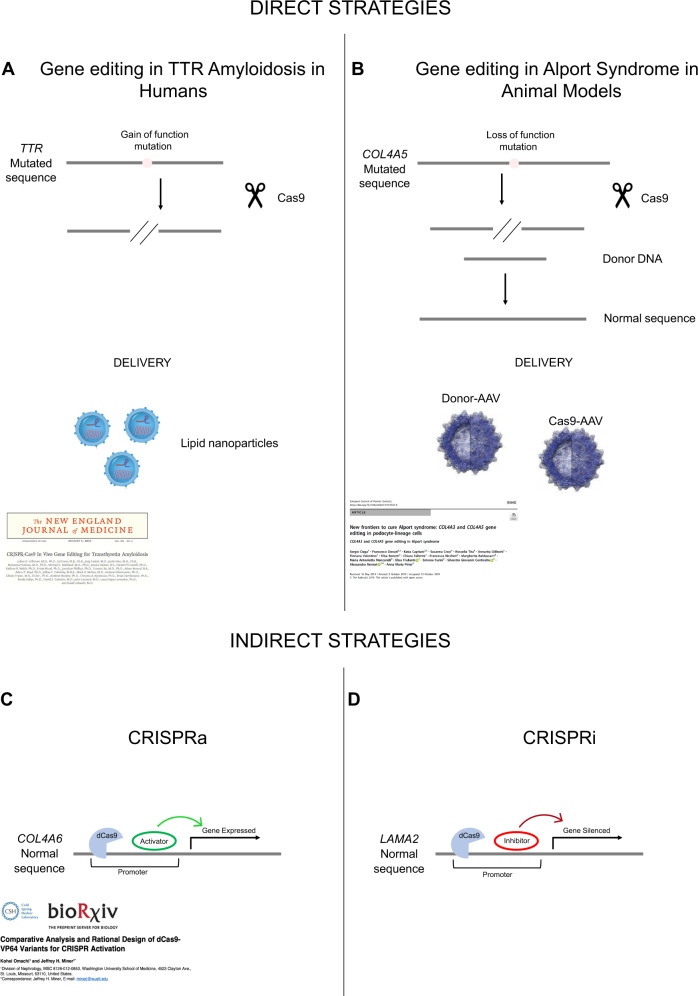
Fig. 3. Gene editing approaches: comparison between direct and indirect strategies.
The CRISPR/Cas9 direct approaches are targeting straightforward mutated genes (A, B). The CRISPR/Cas9 indirect approaches are based on the action of endonuclease deficient activity dCas9 fused to either transcriptional activators or repressors that either upregulate (CRISPRa) or silence (CRISPRi) genes (respectively) relevant to the pathophysiology of AS (C, D). A TTR gene mutated sequence is edited in humans by spCas9 alone. Indel (insertion/deletion) is created after the cut and the gain of function variant (the protein precipitates in aggregates dangerous for the cell) is edited in a loss of function allele. B The loss of function variant in COL4A5 gene is edited to the normal sequence, by means of spCas9, in cultured cells and animal models employing CRISPR/Cas9 approach. C The COL4A6 gene, a minor basement membrane gene, is forced by dCas9/CRISPRa to be overexpressed in podocytes, which should replace the missing α3α4α5(IV) network with the α5α5α6(IV) network; this could slow kidney disease progression. D The LAMA2 downstream gene is silenced by the dCas9/CRISPRi in glomerular cells, which should reduce laminin α2 in the GBM and slow kidney disease progression.