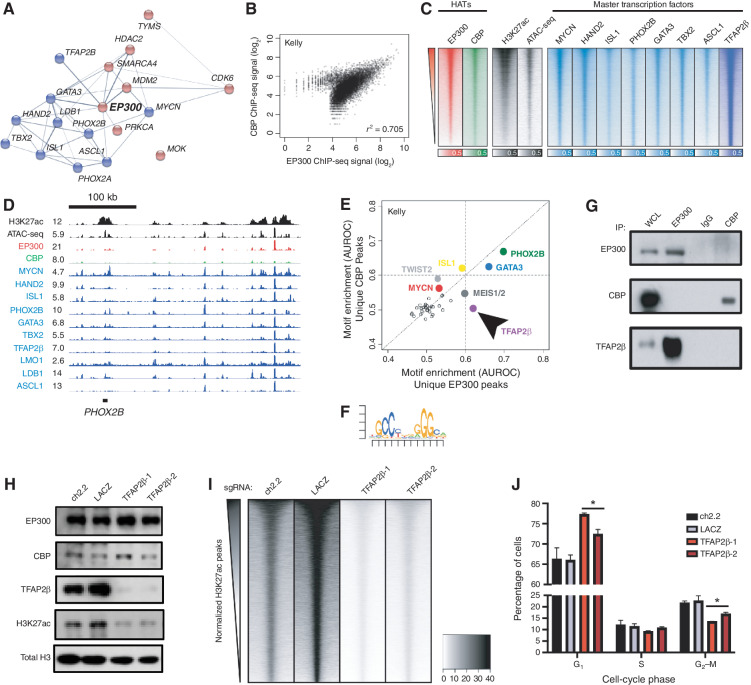
Figure 2.
EP300 regulates the NB CRC directed by TFAP2β. A, STRING database interaction plot of nuclear dependency genes in NB cells. Data are derived from ref. 9. Displayed are CRC TFs in blue and proteins with available targeting compounds in red. Connecting lines indicate previously demonstrated protein–protein interactions. B, Scatter plot of log2-transformed read counts of EP300 or CBP ChIP-seq in the collapsed union of separately identified high-confidence CBP- and EP300-binding sites in Kelly cells. R indicates the Spearman correlation coefficient demonstrating a strong linear relationship in coverage. See Supplementary Fig. S2A for a similar analysis in BE2C cells. C, Genome-wide heat map analysis of chromatin composition at the collapsed union of separately identified high-confidence CRC TF–binding sites in Kelly cells. Rows ordered by EP300 signal. See Supplementary Fig. S2B for a similar analysis in BE2C cells. ATAC-seq, Assay for Transposase-Accessible Chromatin using sequencing. D, Representative ChIP-seq plots demonstrating binding of CRC factors (blue), CBP (green), and EP300 (red) at the PHOX2B CRC TF locus in Kelly NB cells. Also shown is the PHOX2B super-enhancer (H3K27ac) and open chromatin (ATAC-seq, black). Data are representative of both Kelly and BE2C cells and all CRC loci. Values represent normalized reads per million. E, Motif enrichment analysis of ChIP-seq to EP300 and CBP in Kelly NB cells. Data were restricted to the top 500 bound peaks by EP300 or CBP in Kelly NB cells. Colored dots indicate known enriched TFs. Arrow indicates specifically enriched motif, corresponding to TFAP2β. F, Position-weight matrix from analysis in D demonstrates the top enriched specific sequence under EP300 peaks compared with CBP peaks, corresponding to the consensus binding sequence for TFAP2β. G, Co-IP followed by Western blotting analysis of EP300 and CBP in Kelly NB cells. IgG, isotype-matched rabbit IgG antibody; WCL, whole-cell lysate. Data are representative of three independent co-IP Western blots. H, Kelly NB cells expressing Cas9 were infected with sgRNAs targeting TFAP2β (TFAP2β-1,2) or control loci (ch2.2, LACZ), followed by Western blotting to the shown targets. Data are representative of three independent lysates and blots. I, Genome-wide heat map analysis of H3K27ac coverage in wild-type and TFAP2β-knockout Kelly cells using cell number and Escherichia coli spike-in normalized CUT&RUN sequencing. Rows represent 6-kb regions centered on the center of the collapsed union of high-confidence peaks separately identified in each condition and are ordered by control (ch2.2) signal. J, Propodium iodide flow cytometry of Kelly NB cells expressing Cas9 and infected with sgRNAs targeting TFAP2β (TFAP2β-1,2) or control loci (ch2.2, LACZ). n = 3 independent infections and flow analyses. *, P < 0.05. Bars, SEM. See also Supplementary Fig. S2.