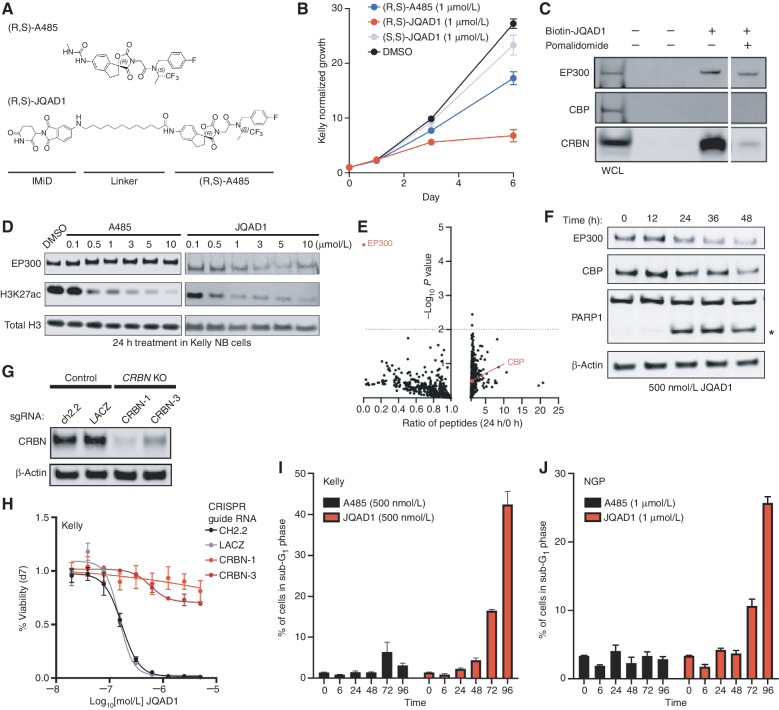
Figure 3.
JQAD1 is a selective EP300 degrader. A, Chemical structure of (R,S)-A485 and (R,S)-JQAD1. B, Kelly cells were treated with 1 μmol/L (R,S)-A485, (R,S)-JQAD1, (S,S)-JQAD1, or DMSO for 6 days, and growth was measured by CellTiter-Glo assay. n = 3 independent experiments and measurements at each time point. Bars, SEM. C, Kelly cell lysates were treated with combinations of Biotin-JQAD1 or pomalidomide prior to streptavidin-bead purification and Western blotting of protein isolates demonstrating enriched interaction of JQAD1 with CRBN and EP300 proteins. WCL, whole-cell lysate. Data are representative of three independent experiments and blots. D, Kelly NB cells were treated with DMSO, A485, or JQAD1 at the noted concentrations (in μmol/L) for 24 hours prior to lysis for Western blotting. Data are representative of three independent biological repeats. E, SILAC-labeled Kelly NB cells were treated with JQAD1 at 500 nmol/L or DMSO vehicle for 24 hours prior to nuclear extraction and analysis by mass spectrometry. Ratio of detected peptides at 0 hours versus 24 hours is demonstrated. Data represent the sum ratio of heavy:light-labeled protein detected in triplicate at 24 hours compared with 0 hours. Dotted line indicates a P value of 0.01. Red labeled points indicate EP300 and CBP. n = 3 independent treatments, lysates, and mass spectrometry reactions. F, Kelly NB cells were treated with JQAD1 at 500 nmol/L for the noted time points prior to lysis for Western blotting. Data are representative of three independent experiments and blots. Asterisk (*) indicates cleaved PARP1 species. G, Kelly cells stably expressing Cas9 were infected with sgRNAs targeting CRBN (CRBN-1,3) or control loci (ch2.2, LACZ), and pools of knockout (KO) cells were established. Western blotting was performed with antibodies against CRBN. Actin is shown as a loading control. Data are representative of three independent Western blots. H, Kelly Cas9 control or CRBN-knockout cells were treated with a range of doses of JQAD1 for seven days, prior to assay by CellTiter-Glo. n = 3 independent replicates per dose and time point. I and J, Propodium iodide flow cytometry of sub-G1 events in Kelly (I) and NGP (J) cells treated with JQAD1 or A485 for the noted time points (in hours). Data are a summary of n > 3 independent flow experiments. Compound treatment was performed at 500 nmol/L (Kelly) and 1 μmol/L (NGP). Similar results were obtained in SIMA cells treated with compounds at 1 μmol/L. Bars, SEM. See also Supplementary Fig. S3.