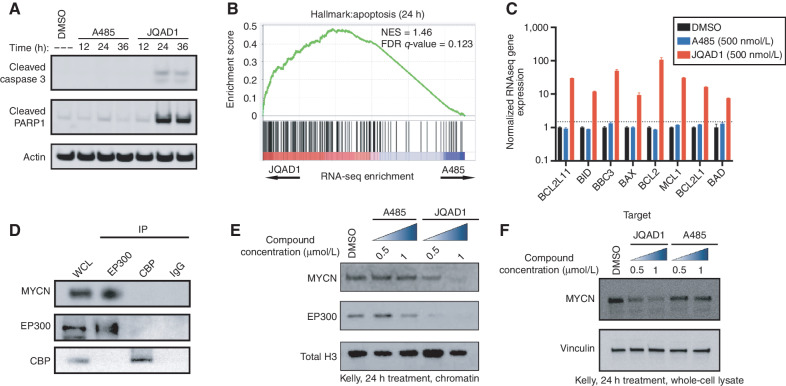
Figure 4.
EP300 degradation rapidly disrupts MYCN expression and causes apoptosis. A, Kelly NB cells were treated with 1 μmol/L JQAD1, A485, or DMSO control for 12, 24, or 36 hours, prior to lysis and Western blotting for the markers of apoptosis: cleaved caspase-3 and cleaved PARP1. Actin is demonstrated as a loading control. Data is representative of three independent treatments and analyses in Kelly and NGP cells. B, Kelly cells were treated with 500 nmol/L JQAD1, A485, or DMSO control for 24 hours prior to External RNA Controls Consortium (ERCC)–controlled spike in RNA-seq. GSEA of RNA-seq results was performed with the MSigDB hallmarks dataset. n = 3 biological replicates and independent RNA extractions per treatment. C, Normalized RNA-seq gene expression of pro- and antiapopotic mRNA transcripts from Kelly cells treated as in B. Log10 transcript expression is shown, normalized against DMSO and ERCC controls. n = 3 biological replicates and independent RNA extractions per treatment. Bars, SEM. D, Nuclear lysates from Kelly cells were immunoprecipitated with anti-EP300, anti-CBP, or IgG control antibodies. WCL, whole-cell lysate. Data is representative of >3 independent co-IP/Western blots. E and F, Kelly cells were treated with DMSO control, A485 (0.5, 1 μmol/L), or JQAD1 (0.5, 1 μmol/L), followed by extraction of chromatin (E) or whole-cell lysates (F) and Western blotting. Total H3 is shown as a loading control. Data are representative of three independent biological replicates. See also Supplementary Fig. S4. NES, normalized enrichment score.