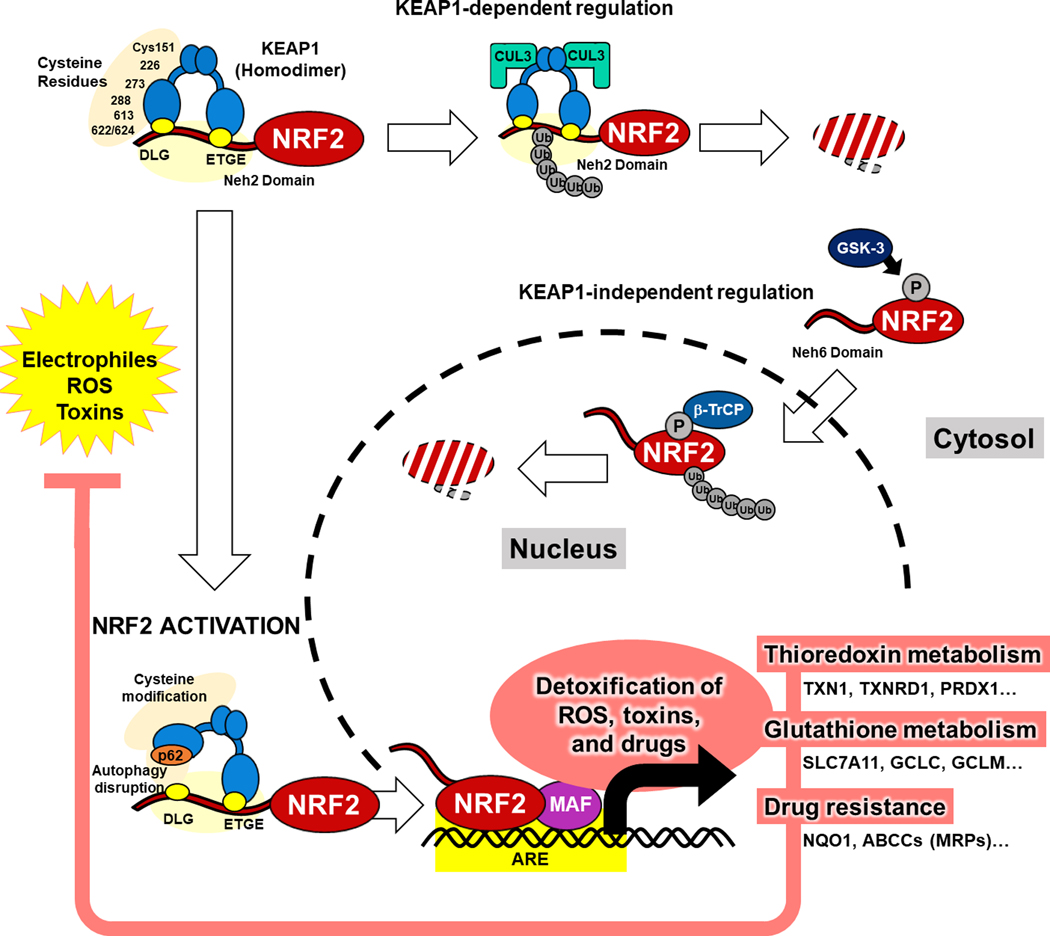
Figure 1: Physiological Activation and Regulation of NRF2.
Under basal conditions, NRF2 is bound by KEAP1 via the DLG and ETGE motifs in the Neh2 domain of NRF2 in cytosol and leads to binding of CUL3, poly-ubiquitination and proteasomal degradation. NRF2 is also regulated by KEAP1-indepent mechanisms via phosphorylation of the Neh6 domain by GSK-3 and proteasomal degradation by β-TrCP. Reactive oxygen species (ROS), drugs, and toxins react with cysteine residues on KEAP1 resulting in structural changes and the accumulation of NRF2 to translocate to the nucleus and function as a transcriptional factor. In the nucleus, NRF2 heterodimerizes with small MAF proteins and binds to antioxidant response elements to induce a series of target genes for detoxification of ROS, toxins, and drugs.
KEAP1, Kelch-like ECH-associated protein 1; NRF2, nuclear factor erythroid 2-related factor 2; β-TrCP, beta-transducin repeat containing protein; GSK-3, glycogen synthase kinase 3; Ub, ubiquitin; ARE, antioxidant response element; MAF, musculoaponeurotic fibrosarcoma; TXN, thioredoxin; TXNRD1, thioredoxin reductase 1; PRDX1, peroxiredoxin-1; GCLC, glutamate-cysteine ligase catalytic subunit; GCLM, glutamate-cysteine ligase modifier subunit; NQO1, NADPH-quinone Dehydrogenase 1; ABC, ATP-binding cassette