Extended Data Fig. 1: Mutual information structure of the TCRβ sequence.
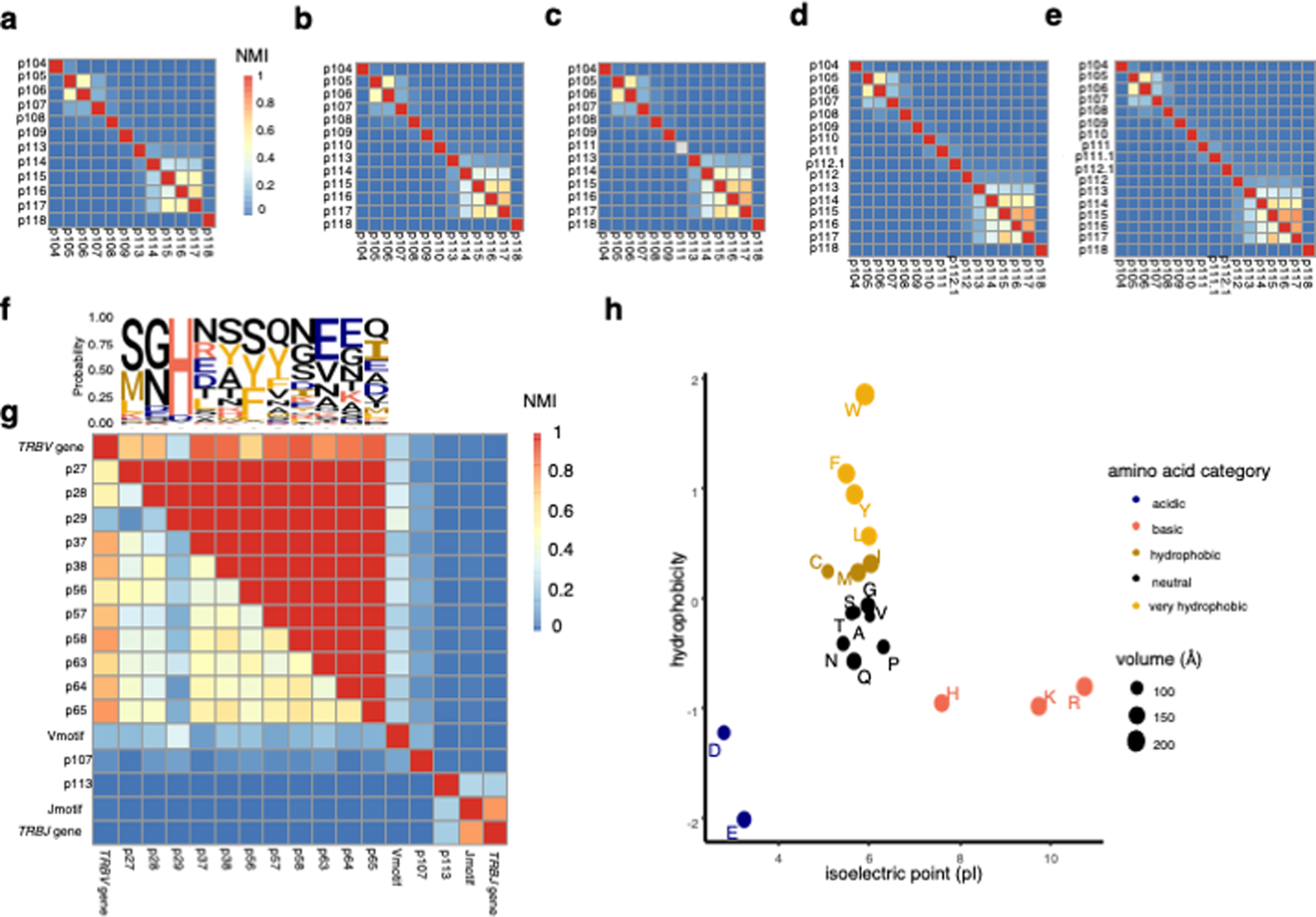
(a) – (e) Heatmap depicting the mutual information structure of the CDR3β amino acid sequence for CDR3βs of length 12 (a), 13 (b), 14 (c), 16 (d), and 17(e) in the discovery dataset. The lower diagonal features normalized mutual information (NMI) between each pair of TCR positions, while the upper diagonal features the maximum mutual information achieved by conditioning on any other TCR position. NMI color scale for (a)-(e) is provided in (a). (f) Probability of each amino acid in each TCR position depicted by a sequence logo. (g) Heatmap as in (a) – (e) for CDR1β and CDR2β loop positions as well as TCR features derived from the flanking regions of CDR3β (Methods). (h) Categorization of amino acids by isoelectric point and interfacial hydrophobicity (Methods).
