Figure 1. PfCRISP shows structural features of SCP family proteins.
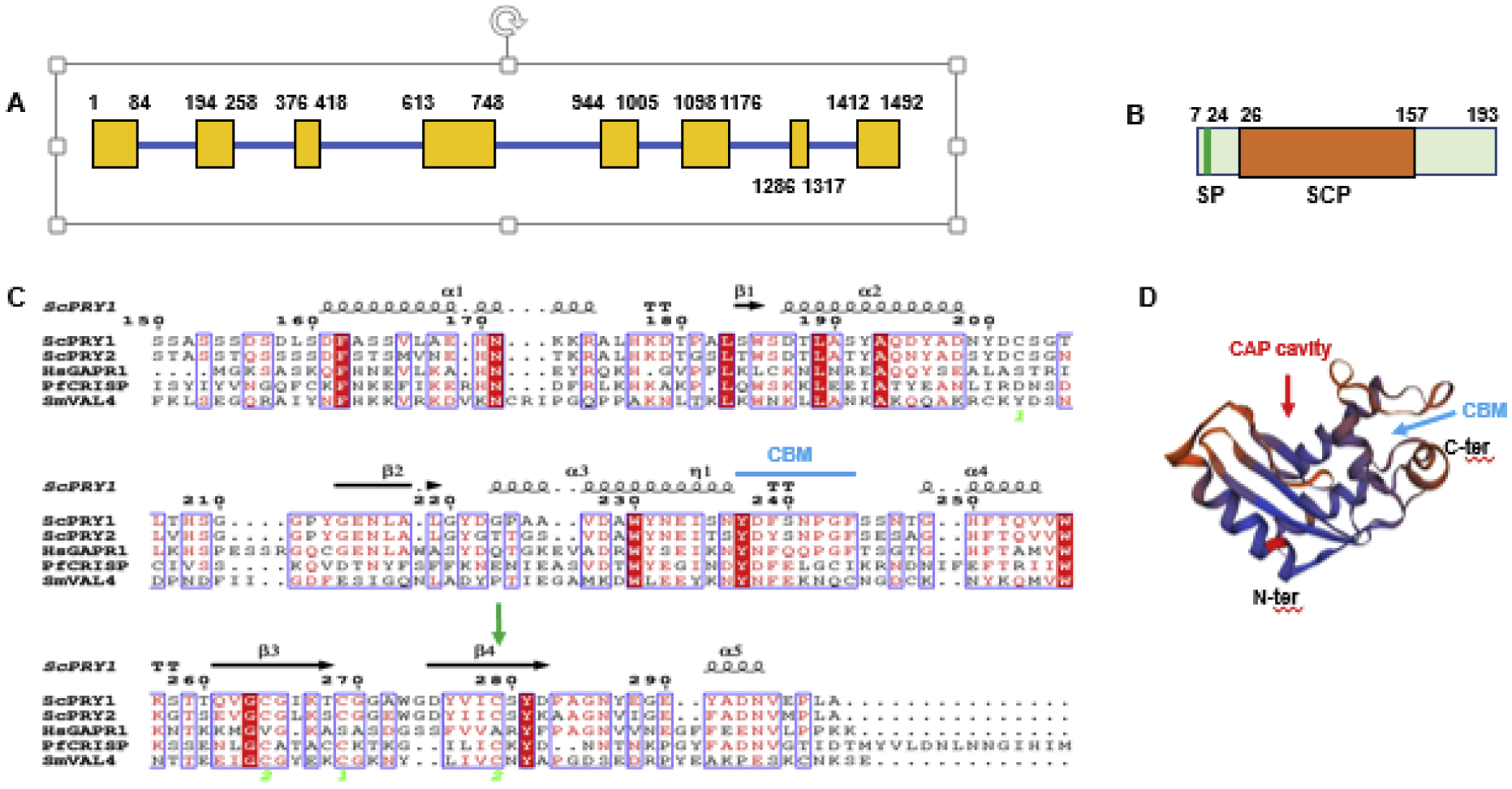
(A) Schematic for the PfCRISP gene showing exons (yellow boxes) and introns (blue bar). (B) PfCRISP domains: SP (signal peptide), in green; SCP domain in orange. (C) Sequence alignment for ScPRY1, ScPRY2, HsGAPR1, PfCRISP and SmVAL4. Conserved residues are in white font on a red background. α-helices and ß sheets are indicated based on structural features of ScPRY1 structure (PDB ID: 5JYS). Cyan arrow indicates CBM (Caveolin Binding motif, FxFxxxxF). Green arrow indicates C147 and its conservation across all the proteins. Sc-Saccharomyces cerevisiae, Hs-Homo sapiens, Pf-Plasmodium falciparum, Sm-Schistosoma mansoni. (D) Predicted three-dimensional structure of PfCRISP. PDB template used 5JYS (ScPRY1). The CAP cavity is marked by red arrow and cyan arrow indicates CBM. See also supplementary Figure S1.
