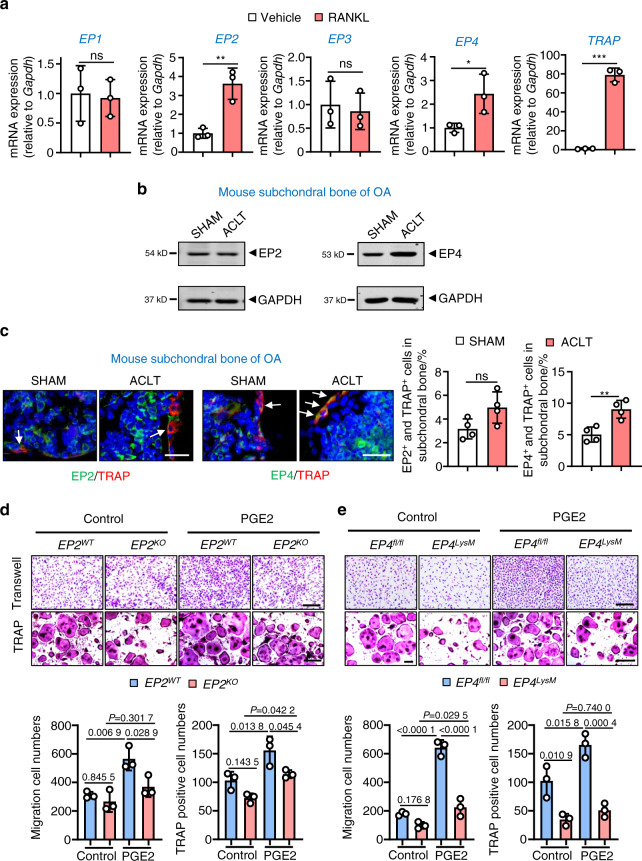
Fig. 1.
Expression of EP2 and EP4 in subchondral bone osteoclasts and the effects of PGE2 on osteoclasts with EP2 and EP4 deletion. a qRT-PCR analysis of the mRNA expression levels of EP1, EP2, EP3, EP4, and Trap in osteoclasts differentiated from BMMs exposed to 10 ng·mL−1 M-CSF and 50 ng·mL−1 RANKL for 5–6 days. Error bars are the mean ± s.d. n = 3. *P < 0.05, **P < 0.01 and ***P < 0.001, ns, not significant by unpaired two-tailed Student’s t test. b Representative images of the protein levels of EP2 and EP4 in mouse tibial subchondral bone tissue 2 weeks after sham or ACLT surgery. The experiments were performed with three biological replicates. c Representative images of IHF staining of EP2 or EP4 (green) and TRAP (red) in subchondral bone 2 weeks post-sham or ACLT surgery (left) and quantitative analysis (right). The white arrows indicate TRAP-positive osteoclasts expressing EP2 or EP4. Error bars are the mean ± s.d. *P < 0.05, ns, not significant by unpaired two-tailed Student’s t test. n = 4 for per group. Scale bars, 20 μm. d EP2 deletion inhibits PGE2-induced migration and osteoclast differentiation. BMMs from the EP2WT and littermate EP2KO mice were used to generate osteoclasts by stimulation with 10 ng·mL−1 M-CSF and 50 ng·mL−1 RANKL and incubation with 100 nmol·L−1 PGE2. Representative image of cells from the Transwell migration assay (Transwell) and osteoclast differentiation assay (TRAP staining) (top) and the corresponding quantitative analysis (bottom). Error bars are the mean ± s.d. n = 3. Two-way ANOVA followed by Tukey’s t tests. Scale bars, 50 μm. e EP2 deletion inhibits PGE2-induced migration and osteoclast differentiation. BMMs from the WT (EP4fl/fl) and littermate EP4fl/fl: LysM-cre (EP4LysM) mice were used to generate osteoclasts by stimulation with 10 ng·mL−1 M-CSF and 50 ng·mL−1 RANKL (5–6 days) and incubation with 100 nmol·L−1 PGE2. Representative image of cells from the Transwell migration assay (Transwell) and osteoclast differentiation assay (TRAP staining) (top) and the corresponding quantitative analysis (bottom). Error bars are the mean ± s.d. n = 3. Two-way ANOVA followed by Tukey’s t tests. Scale bars, 50 μm