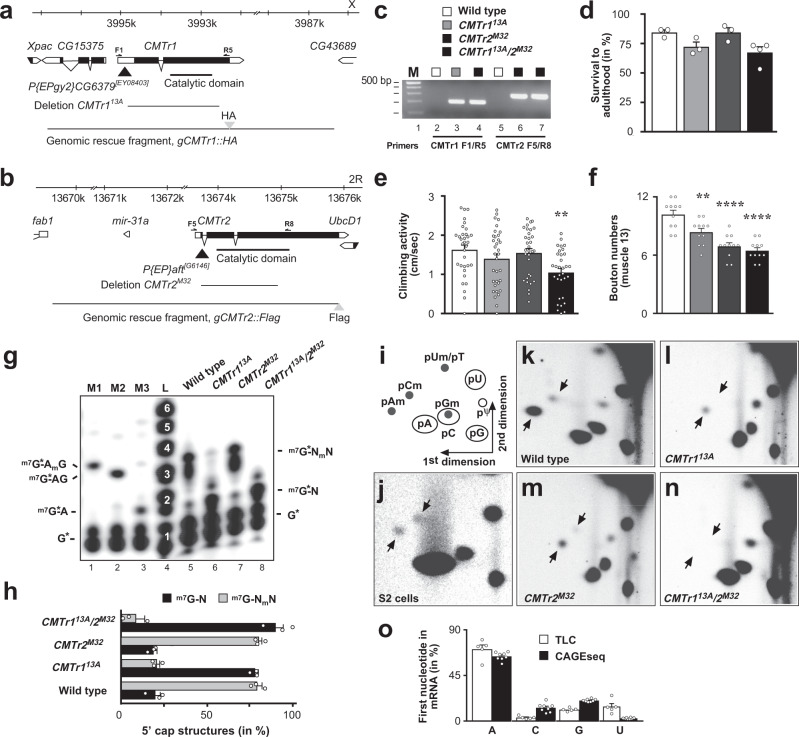
Fig. 1. Analysis of CMTr1 and CMTr2 null mutants and mRNA cap 2′-O-ribose methylation in Drosophila.
a, b Genomic organization of the CMTr1 and CMTr2 loci depicting the transposons (black triangle) used to generate the deletions 13A and M32, which are null alleles. Genomic rescue fragments tagged either with hemaglutinin (HA, a) or FLAG (b) epitopes are indicated at the bottom. Primers used for validating the deletions are indicated on top of the transcript. c Validation of CMTr113A and CMTr2M32 single and double mutants by genomic PCR. The gel is representative of two biological replicates. The marker is a 100 bp DNA ladder with 500 bp indicated on top. Wild type is indicated in white, CMTr113A in light gray, CMTr2M32 in dark gray, and CMTr113A/2M32 in black. d Survival of flies to adulthood after hatching from the eggshell shown as mean ±SE (n = 3, except CMTr113A/2M32 n = 4). e Climbing activity was assessed by negative geotaxis assays shown as mean ± SE, n = 40, p = 0.005 by one-way ANOVA followed by Tukey’s test. f Bouton numbers at NMJs of muscle 13 in third instar larvae are shown as mean ± SE. n = 11, **p = 0.005 and ****p ≤ 0.0001 by one-way ANOVA followed by Tukey’s test. g Recapping of mRNA with 32PalphaGTP from adult flies of the indicated genotypes. 5′cap structures were separated on 20% denaturing polyacrylamide gels after digestion with RNAse I (lanes 5–8, right) Markers—M1: RNAse I digested 32PalphaGTP capped in vitro transcript starting with AGU and 2′-O-ribose methylated with vaccinia CMTr. M2: RNAse T1 digested 32PalphaGTP capped in vitro transcript starting with AGU. M3: RNAse I digested 32PalphaGTP capped in vitro transcript starting with AGU. Sequences of markers are shown on the left and of cap structures from adult flies are shown on the right, N: any nucleotide, *: 32P, m: methyl-group. L: single nucleotide ladder with nucleotide number indicated in white made by alkaline hydrolysis of a 5′ 32P-labeled RNA oligonucleotide. h Quantification of 5′ cap structures shown as mean ± SE. n = 3. Non-ribosemethylated cap is in black and ribose methylated in gray. i Schematic diagram of a 2D thin-layer chromatography (TLC) depicting standard and 2′-O-ribose methylated (m) phospho-nucleotides. ψ: pseudouridine. j–n Representative TLCs from three replicates showing modifications of the first cap-adjacent nucleotides of S2 cells (je), adult control (k), and CMTr113A and CMTr2M32 single (l, m) and double (n) mutant females. o Quantification of the mRNA first nucleotide shown as mean ± SE from TLC (n = 5, white) and CAGEseq data (n = 8, black) from adult Drosophila and S2 cells, respectively. Source data for gels, survival to adulthood, climbing activity, bouton numbers at muscle 13, for 5′cap structures, and first nucleotide in mRNA are provided as a Source Data file.