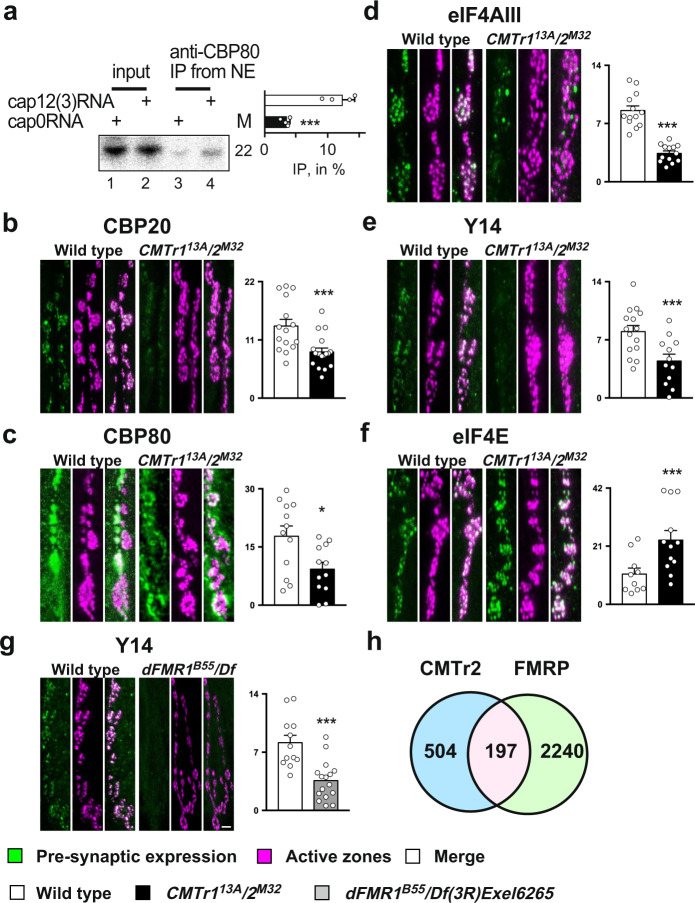
Fig. 5. 2′-O-ribose methylation of mRNA cap-adjacent nucleotides is required for localizing untranslated transcripts to synapses.
a Immunoprecipitation of capped RNA with or without 2′-O-ribose methylation from nuclear extracts. M: length in nucleotides. The graph to the right depicts the ratio of IP/input (n = 4, p = 0.016). b–g Representative images from staining of synapses at third instar NMJs pre-synaptically expressing epitope-tagged markers with elavC155-GAL4 from UAS or stained with an antibody (CBP80) in control (left, green) and CMTr113A; CMTr2M32 double mutant larvae (b–f) or dFMR1B55/Df3R)Exel6265 mutant larvae (g, right, green). The active zone of synapses was stained with nc82 (magenta). The mean ± SE of the intensity is shown on the right is arbitrary units in white for the control, in black for CMTr113A; CMTr2M32 double mutant larvae (b–f) and in gray dFMR1B55/Df3R)Exel6265 mutant larvae (g) for nuclear cap-binding proteins CPB20 (b, n = 16 and 19, ***p = 0.0015) and CBP80 (c, n = 12, *p = 0.05), for nuclear exon junction complex (EJC) proteins eIF4AIII (d, n = 13, ***p = 0.005) and Y14 (e, n = 15 and12, ***p = 0.003), for the rate-limiting translation initiation factor eIF4E (f, n = 10 and 12, ***p = 0.009) and for nuclear exon junction complex (EJC) protein Y14 in control and dFMR1B55/Df3R)Exel6265 mutant larvae (g, n = 12 and 16, ***p = 0.005). The scale bar in g is 1 µm. h Overlap (pink) of CMTr2 CLIP targets (blue) with FMRP targets (green) in Drosophila. Statistical analysis was done by an unpaired t-test. Source data for gel, immunoprecipitations values, and intensity of pre-synaptic bouton stainings are provided as a Source Data file.