FIGURE 5.
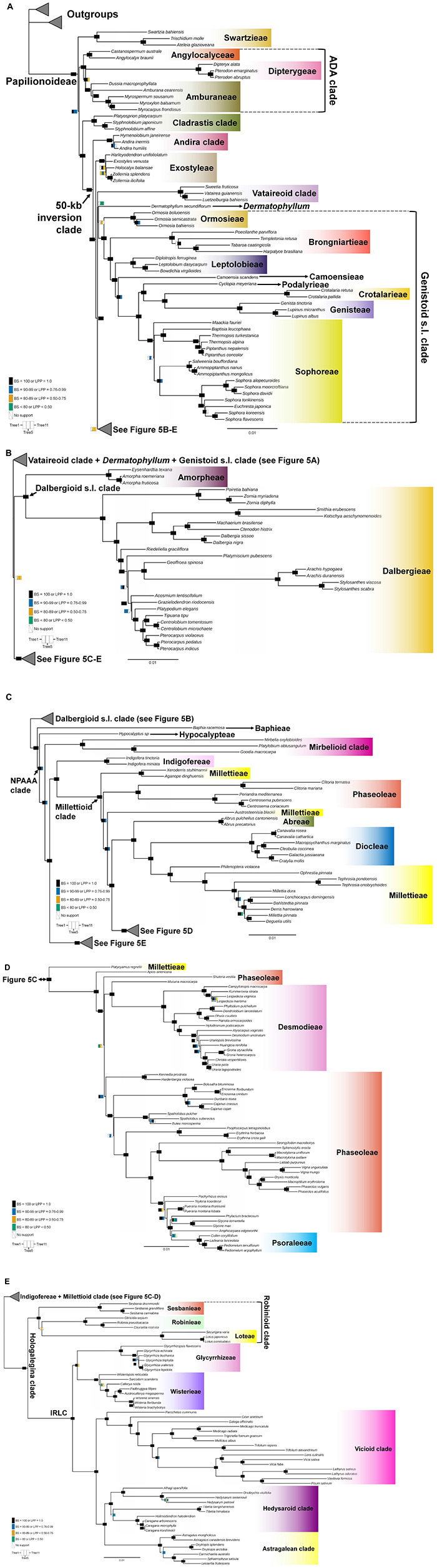
A maximum-likelihood tree based on a concatenated dataset of 77 plastome coding regions with Gblocks trimming (See Table 1). The figure is separated into five panels with a focus on the major clades. (A) Swartzieae, ADA (Angylocalyceae, Dipterygeae, and Amburaneae), Cladrastis, Andira, Exostyleae, Vataireoid, and Genistoid s.l. clades; (B) Dalbergioid s.l. clade; (C) Baphieae, Hypocalypteae, Mirbelioid, Indigofereae, Millettioid (part 1) clades; (D) Millettioid (part 2) clade and (E) Robinioid clade and inverted repeat-lacking clade (IRLC). Supporting values of three trees are visualized at nodes as rectangles. The rectangles are subdivided into three columns that correspond to Tree 1 (left, WP), Tree 5 (middle, CD_nt_gb), and Tree 11 (right, 26 gene trees). Colors in each column represent support values. Black [bootstrap support value (BS) = 100 or local posterior probability (LPP) = 1], Blue [BS (90–99) or LPP (0.76–0.99)], Orange [BS (80–89) or LPP (0.50–0.75)], Bluish green (BS < 80 or LPP 0.50). Scale indicates number of nucleotide substitutions per site. The colors for clades are not related to their support values. We generally followed the color scheme for early-branching papilionoid clades of Figures 2–5 in Cardoso et al. (2013a).
