Figure 2. Directed and undirected graphs and their adjacency matrices.
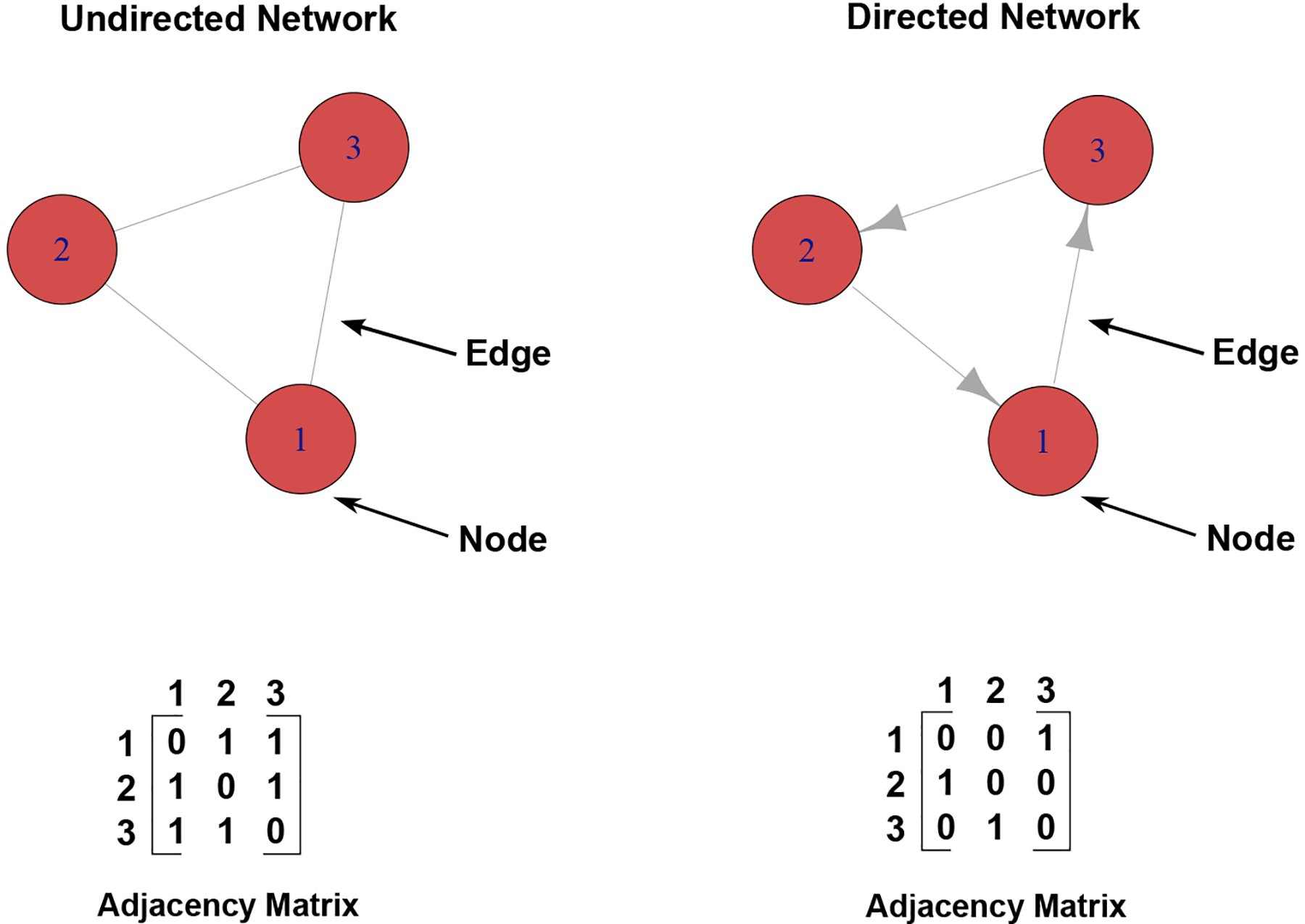
Diagram of the basic structure of an undirected and directed network graph. Each is composed of nodes (which in biological systems generally represent molecules such as genes or proteins) connected by edges (which in biological systems generally represent regulatory or direct functional relationships). Undirected networks only assert that a relationship exists among nodes, and this relationship is presented as symmetric. This feature is reflected in the symmetric adjacency matrix, a matrix representation of the network. In row 1, the given values are 0, 1, and 1, indicating that node 1 is not connected to itself but is connected to nodes 2 and 3. The columns can be read the same way for undirected networks, hence the symmetry of the matrix. Directed networks, by contrast, assert the directionality of the relationship between nodes. In biological networks, this is often intended to indicate that one node is the regulator and the other node is the target. The corresponding adjacency matrix is read slightly differently, where each row indicates the edges going out from that node, while each column represents the edges coming in. Thus, the values 0, 0, 1 in row 1 indicate that node 1 has an edge going into node 3, but not the other two nodes.
