FIGURE 3.
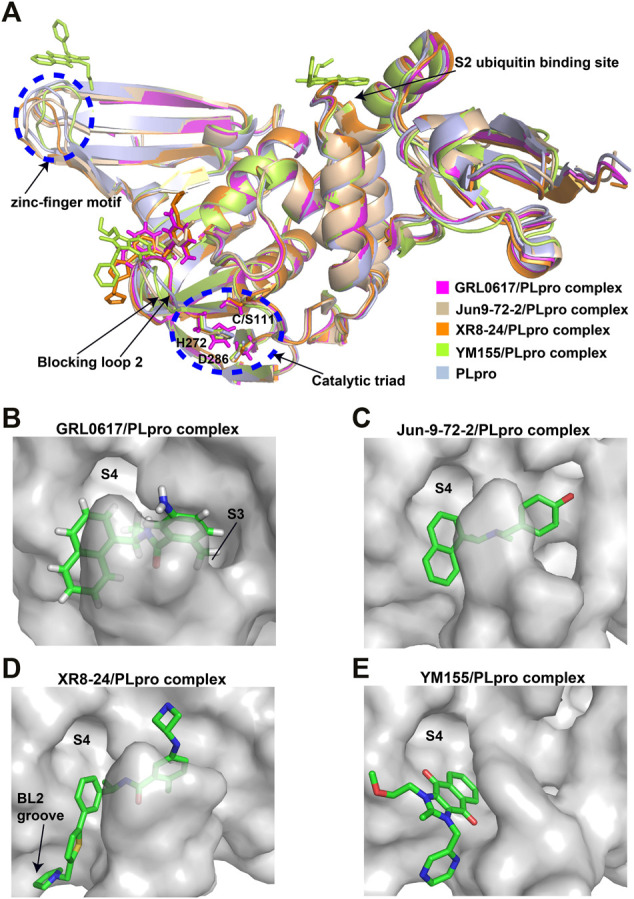
Crystal structure of SARS-CoV-2 PLpro in complex with non-covalent inhibitors. (A) Superposition of PLpro (light blue; PDB ID: 6WZU) with GRL0617/PLpro (magentas; PDB ID: 7CMD), Jun9-72-2/PLpro (wheat; PDB ID: 7SDR), XR8-24/PLpro (orange; PDB ID: 7LBS), and YM155/PLpro (limon; PDB ID: 7D7L) complexes. The catalytic triad and the zinc-finger motif are shown in the blue dashed circle. The blocking loop 2 and the S2 ubiquitin binding site are indicated. (B) Surface presentation of GRL0617 binding pocket. PLpro is shown as gray surface, while GR0617 is shown as sticks. S3 and S4 pockets are labeled. (C) Surface presentation of Jun9-72-2 binding pocket. PLpro is shown as gray surface, while Jun9-72-2 is shown as sticks. S4 pocket is labeled. (D) Surface presentation of XR8-24 binding pocket. PLpro is shown as gray surface, while XR8-24 is shown as sticks. S4 pocket and BL2 groove are labeled. (E) Surface presentation of YM155 binding pocket. PLpro is shown as gray surface, while YM155 is shown as sticks. S4 pocket is labeled.
