Fig. 1 |. Radezolid induces ribosome stalling with alanine in the penultimate position.
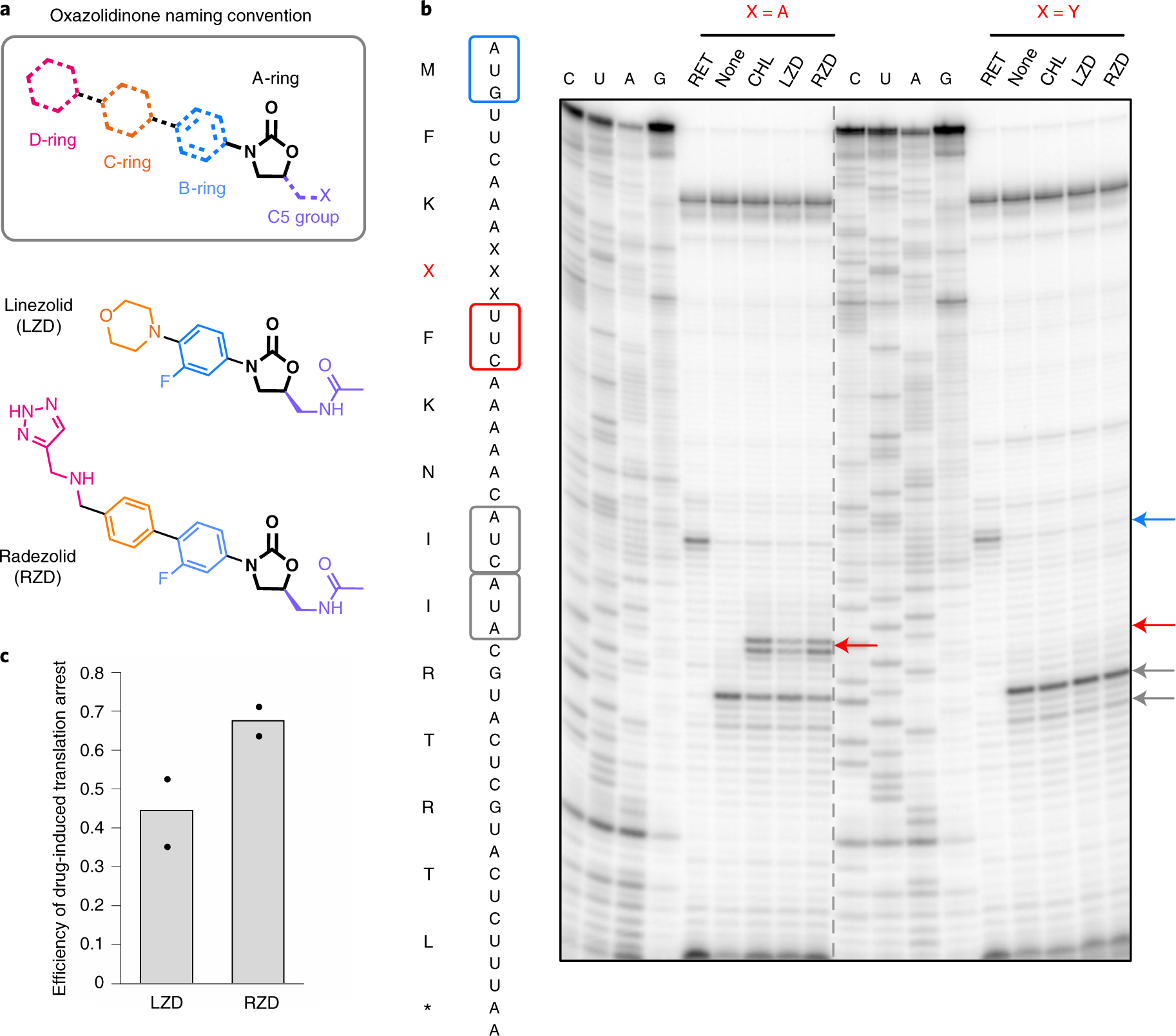
a, Molecular architecture of oxazolidinone antibiotics. The oxazolidinone (A-ring) portion of the molecule is conserved amongst the oxazolidinone class and is shown in black. Chemical moieties that vary among the oxazolidinone derivatives, the C5 moiety and the B, C and D rings are designated by their respective color. b, Toeprinting assays performed on model mRNAs encoding stalling and non-stalling peptides. The control antibiotic retapamulin (RET) was used to stall ribosomes at the start codon, indicated by the blue arrow51. ‘None’ designates reactions lacking ribosome-targeting antibiotics. The control antibiotic CHL was used to stall with alanine at the penultimate position10,11. The toeprint bands corresponding to prominent stall sites observed in reactions containing CHL, LZD or RZD when the fourth amino acid is an alanine, but not tyrosine, are indicated by the red arrow. Owing to the inclusion of the Ile-tRNA synthetase inhibitor mupirocin in all toeprinting reactions, any ribosomes not stalled at an upstream codon are trapped at the downstream Ile codons designated by gray arrows. All antibiotics were added to a final concentration of 50 μM. The presented gel is representative of two independent experiments. c, The extent of translation inhibition by LZD and RZD calculated as the ratio of intensities of the drug-specific toeprint bands (red arrow, b) to the sum of intensities of the drug-specific and trap codon bands (gray arrows, b). The bar graph shows the mean of two independent experiments, with individual data points indicated by black dots.
