Fig. 1.
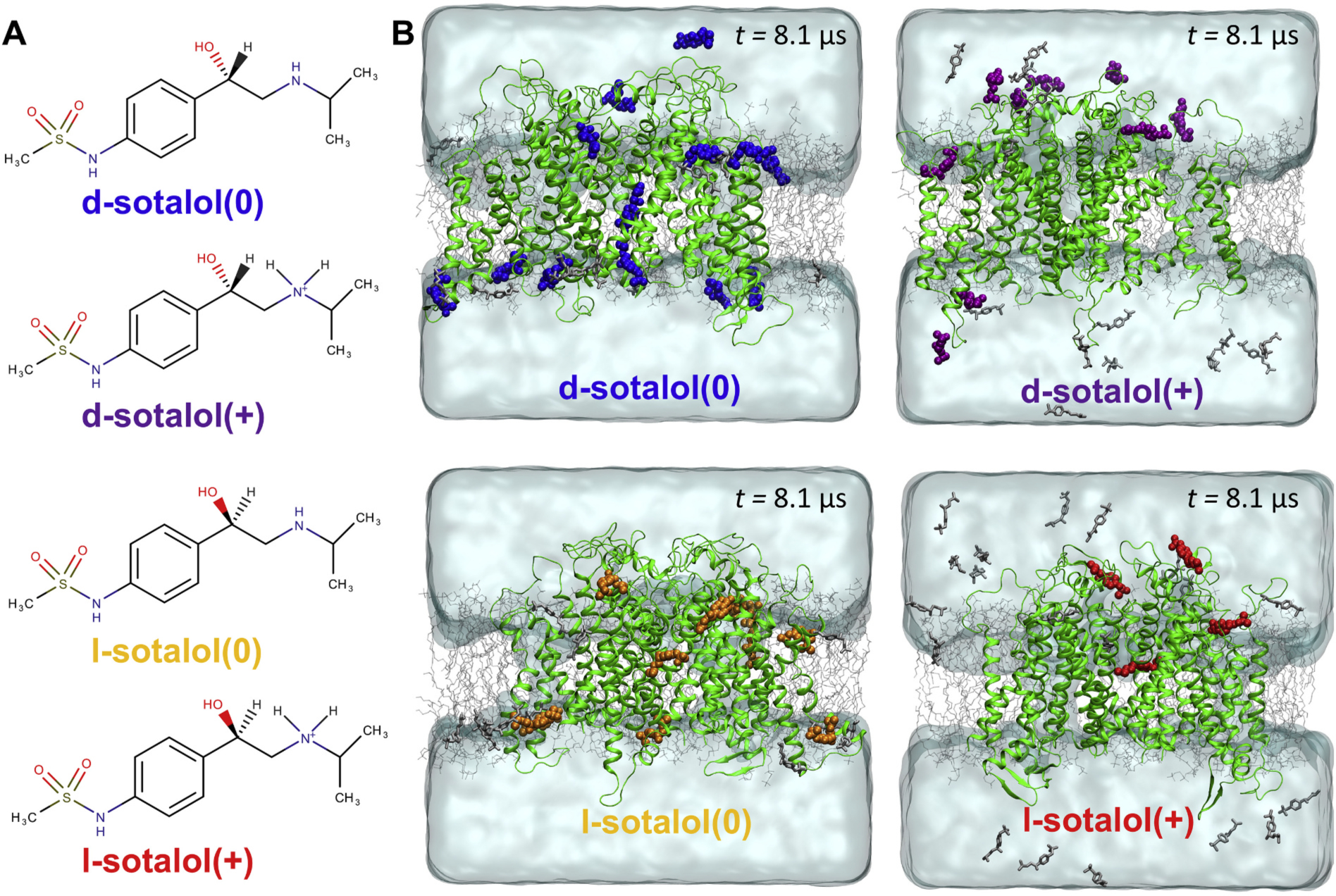
Distribution of cationic(+) or neutral(0) d- or l-sotalol around the hERG channel from multi-μs long unbiased MD simulations. (A) Chemical structures of neutral(0) and cationic(+) forms of d- and l-sotalol (B) Snapshots of the molecular systems consisting of the hERG channel embedded in the POPC bilayer, solvated with aqueous 150 mM KCl and initial 50 mM sotalol solution, at the end of 8.1 μs MD simulations. For sotalol molecules within 3.5 Å of hERG protein residues non‑hydrogen atoms are shown in the colored space filling representation, non-interacting sotalol molecules are shown as gray sticks. The hERG channel is shown as green ribbons, POPC lipid tails as thin gray sticks, water as aquamarine surface, K+ and Cl− ions are not shown for clarity. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
