Figure 2.
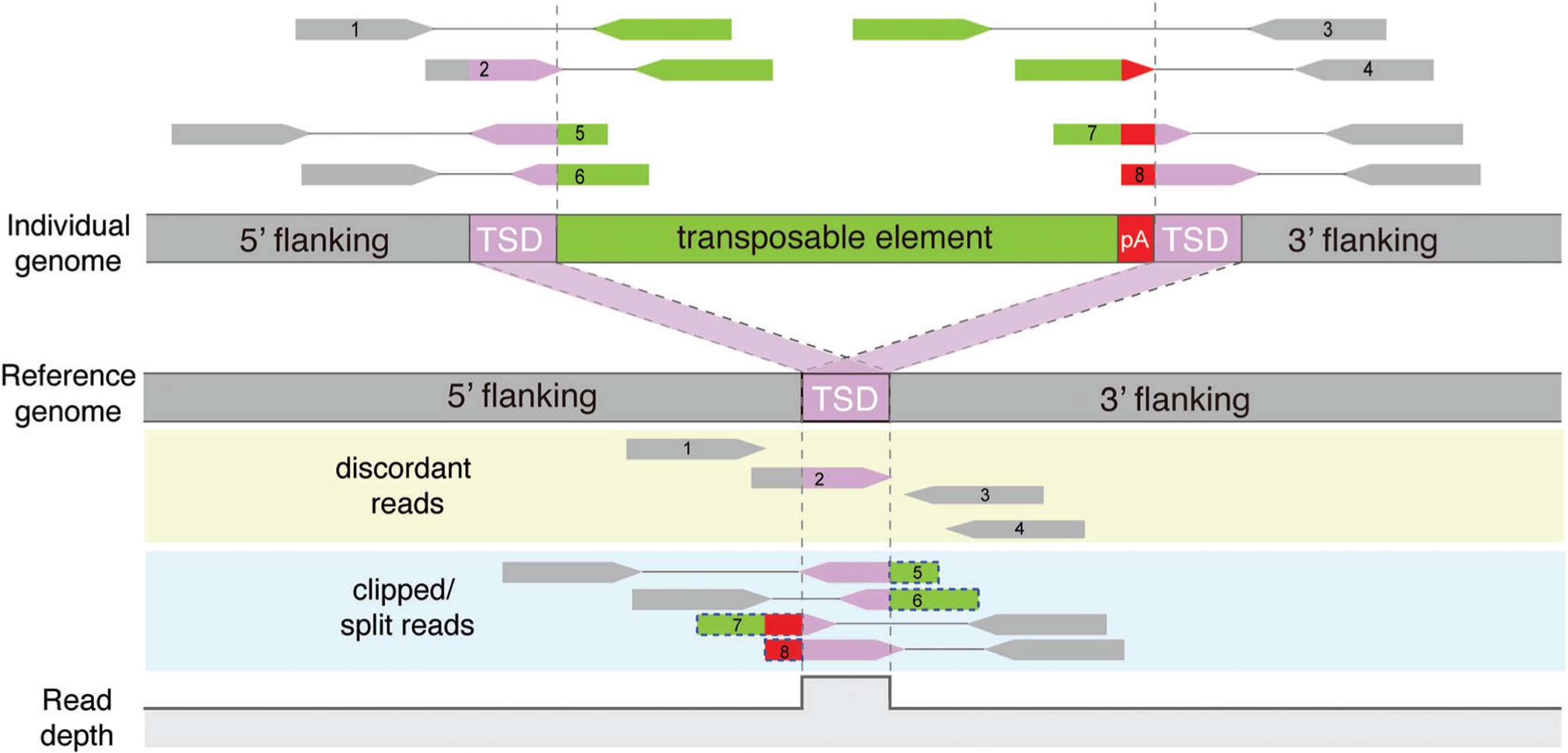
Identification of TE insertions from short-read sequencing data. Paired-end short reads from an individual with a TE insertion are aligned to the reference genome. A TE insertion is detected by identifying two types of read clusters near the insertion breakpoints: (i) discordant reads (reads 1–4) are uniquely aligned to flanking regions and have their mate-pair reads aligned to one of many reference TE copies remotely located from the breakpoints; and (ii) clipped reads or split reads (reads 5–8) span the insertion breakpoints, and thus have soft-clipped or split mapping to the reference (shown in dotted blue boxes). The change in read depth at a non-reference insertion site is shown at the bottom. Gray dashed lines indicate the boundary of TSDs.
