Figure 3.
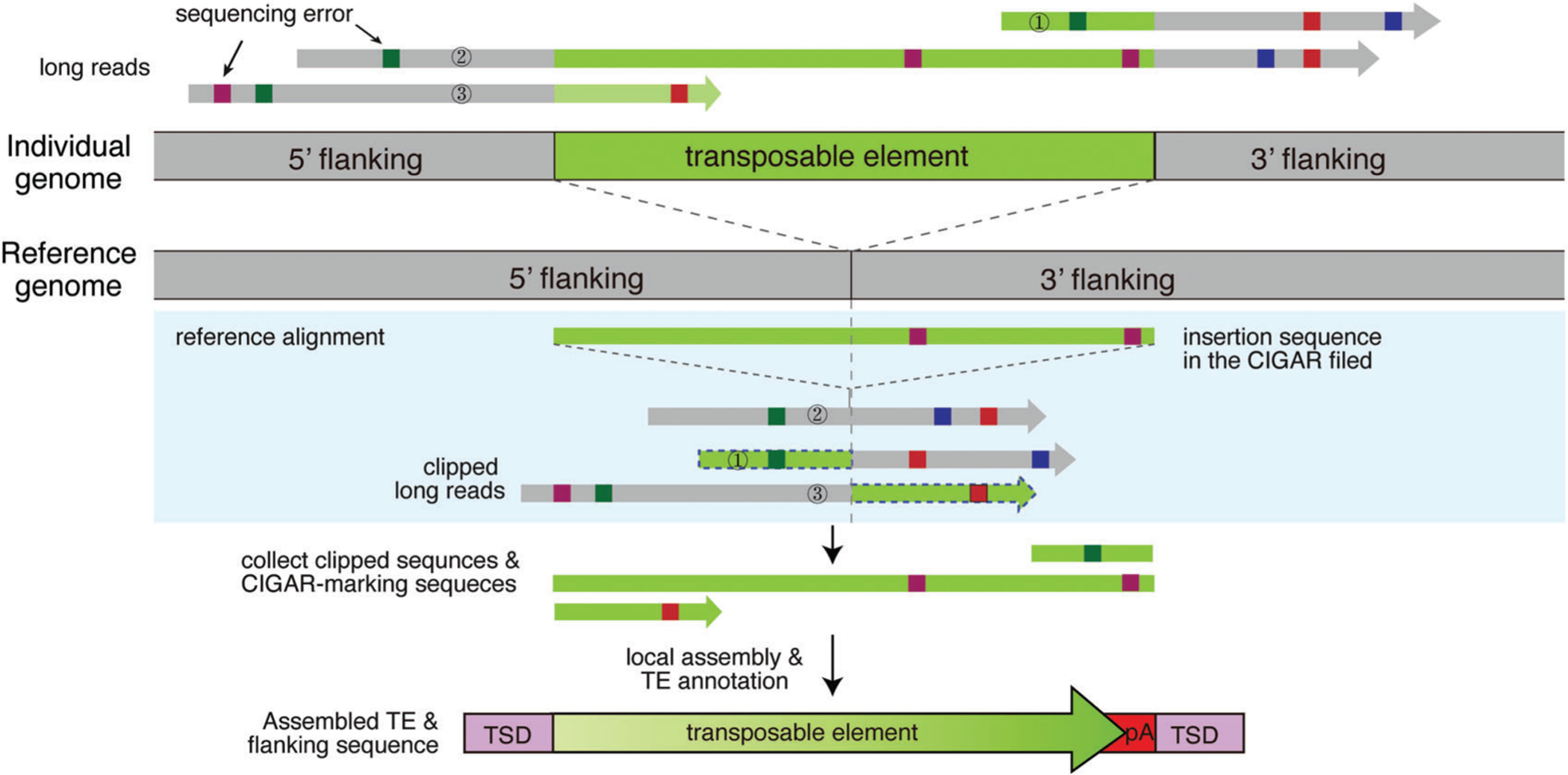
Identification of TE insertions from long-read sequencing data. Long reads from an individual with a TE insertion are aligned to the reference genome. First, insertion-supporting reads are collected: reads with a CIGAR field indicating internal insertions (read 2) and soft-clipped or split reads (read 1 and 3). Local assembly of these insertion-supporting reads is performed to create long contig sequences. The contigs are aligned to TE subfamily sequence libraries, and TE insertions are identified and annotated for multiple features, such as insertion size, TSD, and poly(A) tails.
