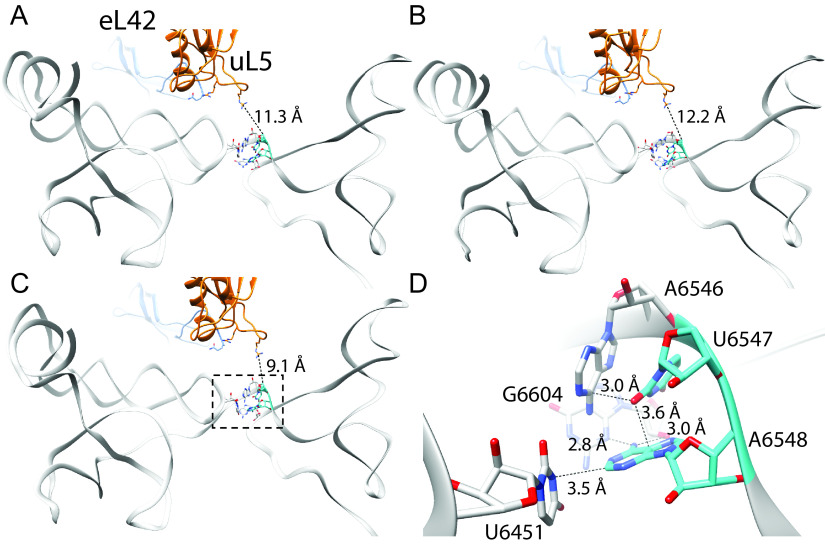
FIG 10.
Hinge residues U6547 and A6548 are important for pretranslocation mimicry by the IAPV IRES. Three conformational states of the pretranslocation IAPV IRES in complex with the 80S ribosome determined by cryo-EM are shown (A–C; PDB 6p5i, 6p5j, and 6p5k respectively) (21). These show small conformational changes in the hinge region and relative orientation with respect to uL5 (blue) and eL42 (orange), which do not form any direct contacts in the pretranslocation state (distance between uL5 Arg58 and U6547 indicated). Close analysis of the hinge region as shown in D (boxed in C) reveals that U6547 adopts a tight “wedge” structure to facilitate the turn in the RNA strand and subsequent positioning of the PKI domain. Both U6547 and A6548 form a specific network of potential hydrogen bond interactions with surrounding nucleotides. Also see Movie S1.