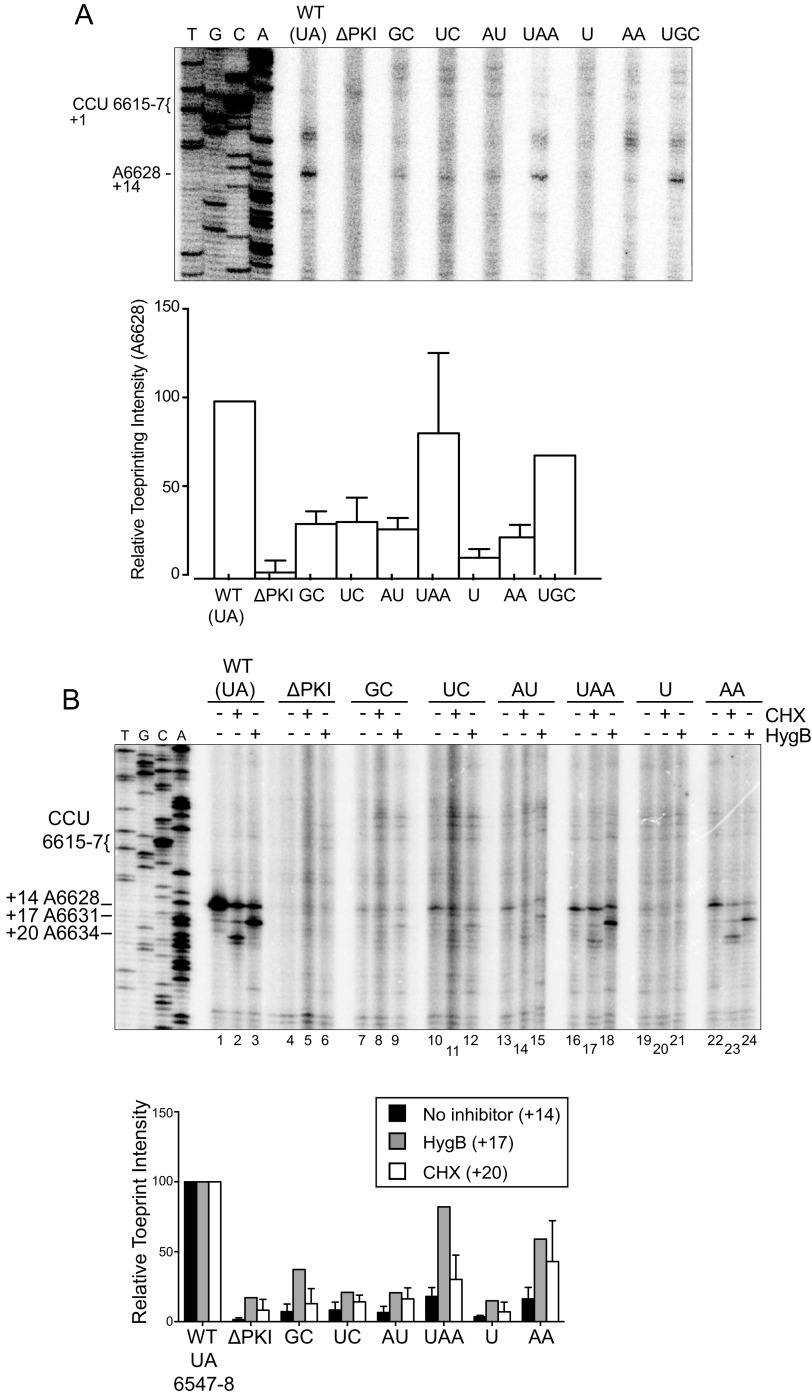
FIG 5.
Toeprinting analysis of IAPV IGR IRES in RRL in the presence of translation inhibitors. Bicistronic RNAs containing the wild-type or hinge mutant IGR IRESs were incubated in absence or presence of (A) lactimidomycin D (LTM 5 μM), (B) cycloheximide (CHX 17.8 μM), or hygromycin B (HygB 1 μM). Primer extension analysis was carried out in the presence of [32P]-dATP, and the reaction products were resolved by denaturing polyacrylamide gel electrophoresis and visualized by autoradiography. (Left) Sequencing ladder for the wild-type IRES is shown with location of the major toeprints. Primary detected toeprints are +14, +17, and +20 nucleotides downstream of the CCU 6615-7 in the ribosomal A-site, given that the first C is +1. A representative gel is shown with the indicated drug treatments. (Below) Quantitation of toeprints. Toeprint intensities were measured as a fraction of the radioactive counts for each toeprint over the total radioactive counts within each lane, normalized to that of the respective toeprints of the wild-type IGR IRES. Averages from at least three independent experiments ± SD are shown for the cycloheximide experiments, and averages from two independent experiments are shown for the hygromycin B experiment.