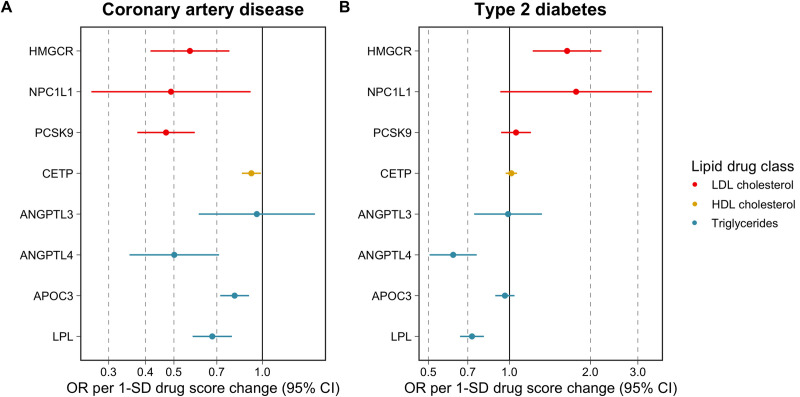
Fig 2. A forest plot visualising the genetically predicted effects of lipid-modifying drug targets on risk of CAD and T2D using MR.
Estimates are colour coded based on the lipoprotein lipid trait estimates used to derive genetic scores. Each genetic score was oriented to mimic the putative effects of drug targets on lipoprotein traits, meaning that effect estimates correspond to relative odds of disease per 1 SD change in either lower LDL cholesterol, higher HDL cholesterol, or lower triglyceride levels via each specific drug target. Note that in the case of CETP, we are not ascribing causal effects to HDL cholesterol—rather, we are orientating CAD/T2D effect estimates corresponding to a genetically predicted increase in HDL cholesterol arising from pharmacological inhibition of CETP. The data underlying this figure can be found in S2 Table. CAD, coronary artery disease; CETP, cholesteryl ester transfer protein; HDL, high-density lipoprotein; LDL, low-density lipoprotein; MR, mendelian randomisation; SD, standard deviation; T2D, type 2 diabetes.