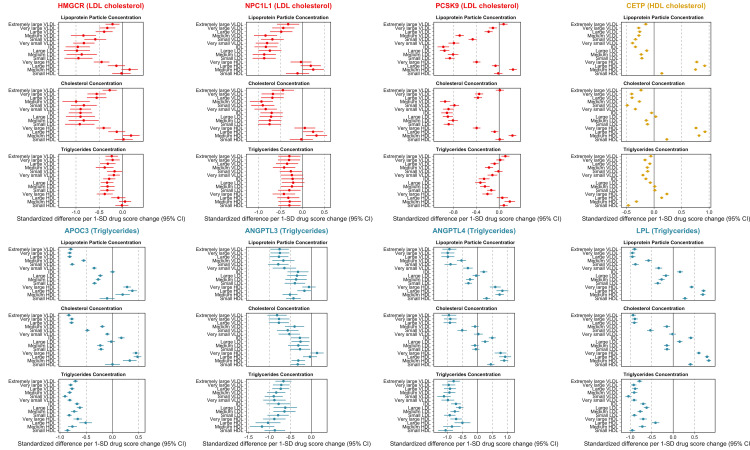
Fig 3. Forest plots illustrating the genetically predicted effects of lipid-modifying drug targets on measures of circulating metabolite concentrations using NMR in the UKB study.
Effect estimates are based on an SD change in the genetically predicted drug target scores oriented to reflect therapeutic intervention (i.e., lower LDL cholesterol, lower triglycerides, and higher HDL cholesterol). Scores were derived using genetic variants robustly associated which lipoprotein lipid traits (as indicated in each target’s legend) at each encoding gene’s region. The data underlying this figure can be found in S7–S14 Tables. CETP, cholesteryl ester transfer protein; HDL, high-density lipoprotein; IDL, intermediate density lipoprotein; LDL, low-density lipoprotein; NMR, nuclear magnetic resonance; SD, standard deviation; UKB, UK Biobank; VLDL, very low-density lipoprotein.