Abstract
The mediobasal hypothalamus (MBH) is the central region in the physiological response to metabolic stress. The FK506-binding protein 51 (FKBP51) is a major modulator of the stress response and has recently emerged as a scaffolder regulating metabolic and autophagy pathways. However, the detailed protein-protein interactions linking FKBP51 to autophagy upon metabolic challenges remain elusive. We performed mass spectrometry–based metabolomics of FKBP51 knockout (KO) cells revealing an increased amino acid and polyamine metabolism. We identified FKBP51 as a central nexus for the recruitment of the LKB1/AMPK complex to WIPI4 and TSC2 to WIPI3, thereby regulating the balance between autophagy and mTOR signaling in response to metabolic challenges. Furthermore, we demonstrated that MBH FKBP51 deletion strongly induces obesity, while its overexpression protects against high-fat diet (HFD)–induced obesity. Our study provides an important novel regulatory function of MBH FKBP51 within the stress-adapted autophagy response to metabolic challenges.
The stress-responsive FKBP51 controls body weight gain by regulating the balance between autophagy and mTOR signaling.
INTRODUCTION
An adequate response to nutritional changes requires a well-coordinated interplay between the central nervous system and multiple peripheral organs and tissues to maintain energy homeostasis. High-caloric food intake and chronic overnutrition strongly challenge this system on a cellular and organismic level and are main drivers in the development of obesity, a hallmark of the metabolic syndrome (1).
Autophagy is an evolutionarily conserved process that efficiently degrades cellular components, like unfolded proteins or organelles, to provide internal nutrients and building blocks for cellular fitness (2). Dysfunctional autophagy is associated with many diseases, such as neurodegeneration, liver disease, cancer, and metabolic syndrome (3–5). In obesity, however, the alterations of autophagy are not fully explored yet. Autophagic signaling in obese individuals is suppressed in pancreatic B cells, liver, and muscle, whereas other studies demonstrated enhanced autophagy signaling in adipose tissue (3, 6, 7). Autophagy initiation is tightly controlled by a series of proteins encoded by autophagy-related genes (ATGs) and regulatory heteroprotein complexes, including the systemic energy sensor adenosine 5′-monophosphate (AMP)–activated protein kinase (AMPK) that is in balance with the mechanistic target of rapamycin (mTOR) (8). Amino acid surplus activates mTOR signaling, which further suppresses autophagy by inhibiting the UNC51-like kinase 1 (ULK1) complex [composed of ULK1, ATG13, FIP200 (FAK family kinase-interacting protein of 200 kDa), and ATG101]. In the course of nutrient deprivation, elevated AMP activates AMPK to initiate autophagy via the ULK1 complex and in turn diminishes mTOR signaling (9, 10). It has recently been shown that the tryptophan-aspartic acid (WD)–repeat proteins that interact with the phosphoinositide protein family (WIPI proteins) act as subordinate scaffolders of the liver kinase B1 (LKB1)/AMPK/TSC2 (tuberous sclerosis complex 2)/FIP200 network linking AMPK and mTOR signaling to the control of autophagy upon metabolic stress (11).
In the mediobasal hypothalamus (MBH), the brain’s central region for metabolic control, the deletion of ATG7 (a ubiquitin E1-like ligase, downstream of the ULK1 complex) in proopiomelanocortin (POMC)–expressing neurons resulted in obesity and dampened sympathetic outflow to white adipose tissue (WAT) (12), while ATG7 deficiency exclusively in agouti-related protein (AgRP)–expressing neurons resulted in decreased body weight (13). Together, these data indicate a role of MBH autophagy in the development of obesity, but key regulatory proteins remain largely elusive.
The FK506-binding protein 51 (FKBP51, encoded by Fkbp5) is the main modulator of the stress response and is best characterized as a co-chaperone to HSP90, thereby orchestrating diverse pathways important to maintain homeostatic control (14–17). We and others have provided evidence that FKBP51 is associated with type 2 diabetes and have shed light on its role as a fundamental regulator of obesity and glucose metabolism (18–22). Following the identification of FKBP51 as a negative regulator of the serine/threonine kinase AKT in cancer cells (23), we showed that FKBP51 acts as a modulator of glucose uptake by mediating the AKT2/PHLPP (PH domain leucine-rich repeat-containing protein phosphatase)/AS160 (AKT substrate of 160 kDa) complex specifically in muscle (18). Furthermore, we demonstrated that FKBP51 induces autophagy through autophagy-promoting beclin-1 (BECN1) in two ways: (i) FKBP51 limits AKT-directed inhibitory phosphorylation of BECN1 at S234 and S295 (24, 25), and (ii) it reduces AKT-mediated phosphorylation of S-phase kinase-associated protein 2 (SKP2) at S72 and thereby lowering its E3-ligase activity, preventing BECN1 from proteasomal degradation (26). Autophagy and FKBP51 are involved in the regulatory role of adipocyte differentiation and mass development (19, 27–29) and are up-regulated in the MBH following starvation (13, 30, 31). The data indicate converging mechanisms of FKBP51-directed protein scaffolding and autophagy, and therefore, we hypothesized a subordinate role of FKBP51 in concert with members of the autophagy signaling network in shaping central and peripheral autophagy signaling. In the current study, we set out to identify the molecular interplay of FKBP51 with cellular autophagic signaling and tested whether FKBP51 shapes the in vivo whole-body response to an obesogenic challenge. Our study unravels the tissue specificity of autophagy signaling in response to obesity and reveals FKBP51 as a previously unknown regulatory link between the stress-induced LKB1/AMPK-mediated autophagy induction and WIPI protein scaffolds.
RESULTS
FKBP51 deletion increases AMPK and mTOR-associated amino acid and polyamine biosynthesis
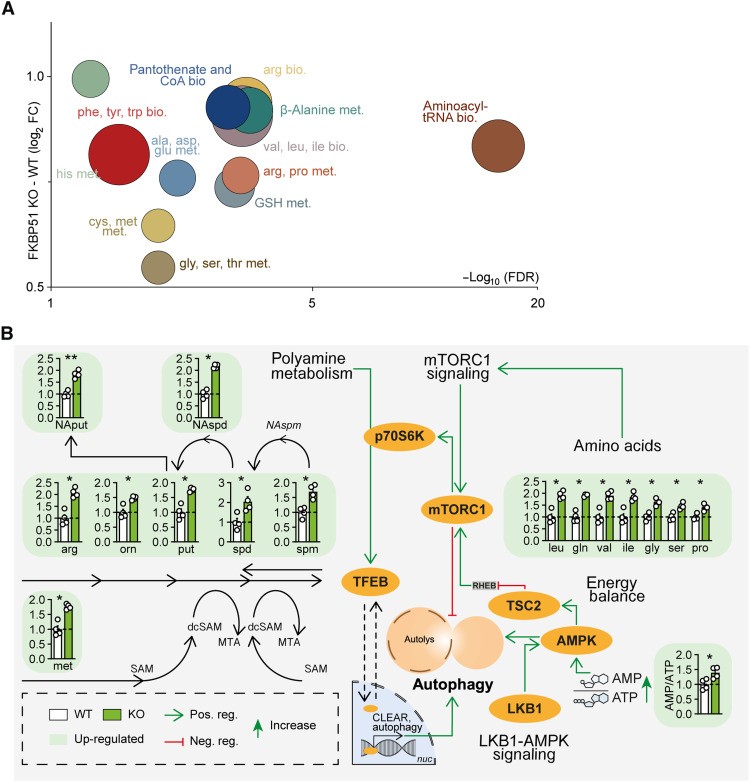
As a first approach to broaden our insights into the contribution of FKBP51 under basal (1× glucose) and metabolically challenging (2× glucose) conditions, we performed a multilevel mass spectrometry (MS)–based metabolomics profiling analysis of human neuroblastoma SH-SY5Y cells lacking FKBP51 [(FKBP51 knockout (KO)] and wild-type (WT) controls. FKBP51 KO cells showed an increase in multiple metabolites compared to WT cells to different extents under basal and high glucose conditions (fig. S1, A to D). Most frequent and pronounced alterations could be attributed to biosynthetic and metabolic pathways of various amino acids, including but not limited to arginine, valine, leucine, and isoleucine biosynthesis and histidine, cysteine, and methionine metabolism (Fig. 1A and fig. S1E). It is well known that amino acids signal to mTOR and that mTOR itself actively participates in the sensing of amino acids in the lysosomal lumen. Particularly, the branched-chain amino acids (BCAAs) leucine, valine, and isoleucine, all elevated in FKBP51 KO cells (Fig. 1B, top right), can activate mTOR and thereby block the autophagy pathway (32–34). On the other hand, the pathway analysis showed increased AMP/adenosine 5′-triphosphate (ATP) ratio and strongly increased levels of polyamines and their metabolites in FKBP51 KO cells (Fig. 1B, left and bottom right). AMP/ATP ratio was strongly reduced in FKBP51 KO cells under metabolically challenging conditions (fig. S2E).
Fig. 1. FKBP51 associates with amino acids and polyamine biosynthesis pathways.
(A) Analysis and regulation of significantly altered pathways of FKBP51 KO and WT cells. The f(x) axis shows the (median) log2 fold change (FC) of all significantly altered metabolites of the indicated pathway, and the false discovery rate (FDR) (equals the −log10–adjusted P value) is shown on the x axis. The size of the circles represents the amount of significantly changed metabolites in comparison to all metabolites of a particular pathway. tRNA, transfer RNA. (B) FKBP51 deletion increases metabolites of the polyamine pathway, the AMP/ATP ratio, and enhances levels of amino acids associated with mTOR signaling. Data in (B) are shown as means + SEM and were analyzed by a two-way analysis of variance (ANOVA) and a subsequent Bonferroni multiple comparison analysis. ala, alanine; AMPK, AMP-activated protein kinase; arg, arginine; asp, asparagine; bio., biosynthesis; CoA, coenzyme A; cys, cysteine; dcSAM, decarboxylated S-adenosylmethione; gln, glutamine; glu, glutamic acid; gly, glycine; GSH, glutathione (reduced); his, histidine; ile, isoleucine; leu, leucine; LKB1, liver kinase B1; met, methionine; met., metabolism; MTA, 5′-methylthioadenosine; mTORC1, mechanistic target of rapamycin complex 1; NAput, N-acetylputrescine; NAspd, N-acetylspermidine; NAspm, N- acetylspermine; orn, ornithine pro, proline; phe, phenylalanine; put, putrescine; SAM, S- adenosylmethionine; ser, serine; spd, spermidine; spm, spermine; TFEB, transcription factor EB; TSC2, tuberous sclerosis complex 2; thr, threonine; trp, tryptophan; tyr, tyrosine; val, valine. *P < 0.05, **P < 0.01.
The overall increase in amino acids most likely results from ubiquitous protein degradation, in line with enhanced cellular catabolic processes. We performed isotope tracing with 13C-labeled glucose to determine the intracellular flux in FKBP51 KO cells. Here, we observed no substantial differences between both cell types (fig. S1F), corroborating the fact that cellular catabolic processes rather dominate over anabolic processes. To exclude that the observed effects are specific to a human cell line, we performed the metabolomics profiling also in mouse Neuro2a (N2a) cells (fig. S2, A to E). While the effects of amino acid biosynthesis are more pronounced in the human cell line following FKBP51 deletion, the data nonetheless point in the same direction and support our hypothesis that FKBP51 affects autophagy and mTOR signaling. We were therefore further encouraged to disentangle the underlining metabolic pathways.
FKBP51 is essential for homeostatic autophagy following nutrient deprivation
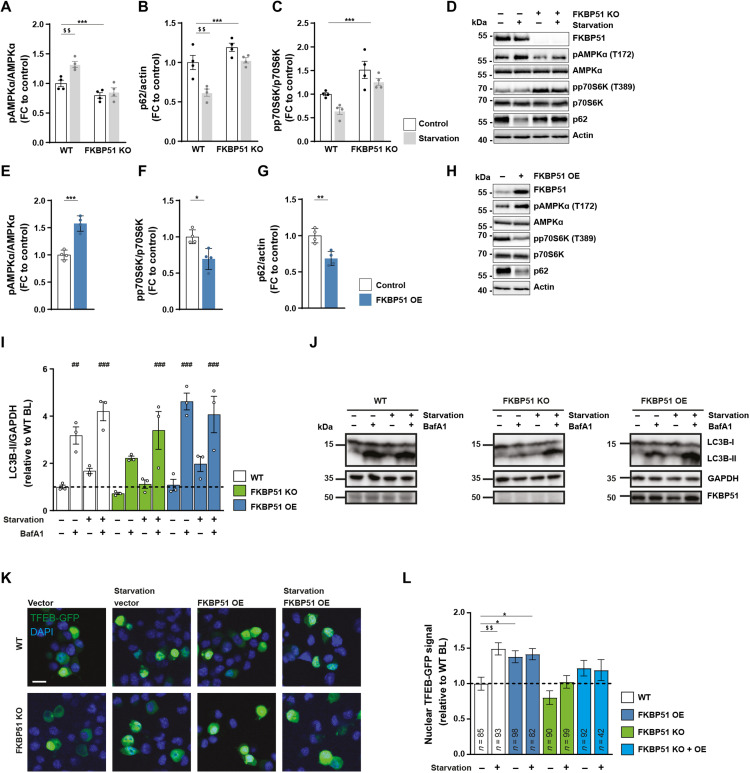
Because autophagy and FKBP51 expression levels are highly induced after starvation (4, 31), we tested whether deletion of FKBP51, in turn, affects the induction of autophagy after nutrient deprivation. To do so, we exposed FKBP51 KO or WT cells to Hank’s balanced salt solution (HBSS) for 4 hours to induce cellular starvation. FKBP51 KO cells showed less phosphorylated AMPK (pAMPK) at T172 and lower levels of LKB1 compared to WT cells already under nutrient-rich conditions (Fig. 2A and fig. S3A), while there was a significant increase in activating phosphorylation of SKP2 at S72 and AKT at S473 (fig. S3, B and C). These changes in upstream signaling resulted in the slight but not significant accumulation of the autophagy receptor and substrate p62 (Fig. 2B), which is an important measure to determine autophagic activity (35). On the other hand, we could observe increased levels of pp70S6K at T389 in FKBP51 KO cells (Fig. 2C). These data imply a reduction of autophagy signaling and increased mTOR signaling after FKBP51 deletion under basal conditions (36). The deletion of FKBP51 blocked the starvation-induced increase in LKB1 protein and the activation of AMPK at T172 (Fig. 2A and fig. S3A) and thereby reduced the level of autophagy signaling. These data underline the importance of FKBP51 in the autophagic stress response after starvation and suggest a tight regulation of FKBP51 on AMPK and LKB1.
Fig. 2. FKBP51 regulates AMPK and mTOR activity following nutrient deprivation.
(A) WT or FKBP51 KO cells were starved in HBSS medium for 4 hours to induce autophagy, followed by quantification of pAMPKα (T172), (B) p62, and (C) pp70S6K (T389). Representative blots are shown in (D). FKBP51 overexpression (FKBP51 OE) in N2a cells (see fig. S3D for validation) enhanced autophagy signaling. Quantification of (E) pAMPKα (T172), (F) pp70S6K (T389), (G) p62, and (H) representative blots. (I) Quantification of autophagic flux in FKBP51 KO and FKBP51 OE cells in response to starvation. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (J) Representative blots of autophagic flux measurements. (K) Representative pictures of TFEB nuclear localization/translocation. DAPI, 4′,6-diamidino-2-phenylindole. Scale bar, 10 μm. (L) Quantification of TFEB reporter assay. BL, baseline. All data (A to J) are shown as relative fold change compared to control condition; ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001; ##P < 0.01, ###P < 0.001; $$P < 0.01. Two-way ANOVA was performed in (A) to (C) and followed by a Tukey’s multiple comparisons test. One-way ANOVA was performed for (I) and (L), followed by a Dunnett’s multiple comparison test. The unpaired Student’s t test was performed for (E) to (G). *, significant genotype effect; $, significant starvation effect; #, significant treatment effect.
Next, we investigated whether FKBP51 up-regulation can enhance autophagy by moderately overexpressing (OE) Flag-tagged FKBP51 in N2a cells (fig. S3D). In line with our hypothesis, FKBP51 OE resulted in highly increased phosphorylation of AMPK at T172 and enhanced levels of LKB1 (Fig. 2E and fig. S3E), which indicate an increase of upstream autophagy initiation, further evidenced by increased phosphorylation of ULK1 at S555 (fig. S3E). Consequently, there was an increase in proautophagic phosphorylation of BECN1 at S14 and S91/S94 (in humans at S93/S96) through kinases ULK1 and AMPK, respectively (fig. S3E) (37). Previous literature has shown that increased LKB1/AMPK signaling also activates the TSC1/TSC2 complex, which, in turn, inhibits mTOR activity (38). In line with that, phosphorylation of TSC2 at S1387 was significantly enhanced (fig. S3E), and the phosphorylation of AKT at S473, SKP2 at S72, and the mTOR substrate pp70S6K at T389 was significantly decreased (Fig. 2F and fig. S2F). In addition, we validated autophagy signaling by the increase in phosphorylation of ATG16L1 at S278, a recently described marker for autophagy induction (39), and the decrease in the autophagy substrate p62 (Fig. 2G and fig. S2G). Last, we assessed autophagic flux by the quantification of LC3B-II (lipidated microtubule-associated proteins 1A/1B light chain 3B) accumulation in response to starvation (HBSS) and the autophagy inhibitor bafilomycin A1 (BafA1) in FKBP51 OE, KO, and FKBP51 KO + OE N2a cells (Fig. 2, I and J, and fig. S3, H and I). BafA1 led to a significant increase in LC3B-II levels in FKBP51 WT and FKBP51 OE N2a cells, but not in FKBP51 KO and FKBP51 KO + OE N2a cells, as compared to FKBP51 WT N2a cells during baseline conditions. Under starvation conditions (HBSS treatment), BafA1 caused a significant accumulation of LC3B-II in all genotypes; however, this increase was attenuated in FKBP51 KO N2a cells.
The data from our metabolomic analysis indicated that alteration in FKBP51 levels might enhance hypusination of the translation factor elF5A and thereby positively regulate autophagy via nuclear translocation of transcription factor EB (TFEB) (40). To test this hypothesis, we measured the translocation of TFEB into the nucleus using a TFEB–green fluorescent protein (GFP) reporter assay in N2a cells. Here, we observed reduced baseline levels of nuclear TFEB in FKBP51 KO N2a cells and diminished nuclear translocation of TFEB after starvation (HBSS) of FKBP51 KO cells compared to the control (Fig. 2, K and L). OE of FKBP51 in WT cells could further increase nuclear TFEB under basal conditions. The nuclear TFEB signal could not be further increased by OE under starvation compared to the WT control (Fig. 2, K and L). We could not detect any differences in the hypusination of eIF5A (fig. S4, A and B).
Together, these findings demonstrate that FKBP51 regulates autophagy induction, especially after a metabolic challenge. However, it is still unclear whether FKBP51 shapes the autophagic response via direct protein-protein interactions to AMPK and LKB1. Given the fact that FKBP51 was previously shown to interact with several signaling molecules within the autophagy pathway (Fig. 3A), we were encouraged to unravel the underlying molecular mechanism and identify potential novel interactions of FKBP51 with members of the autophagy signaling network.
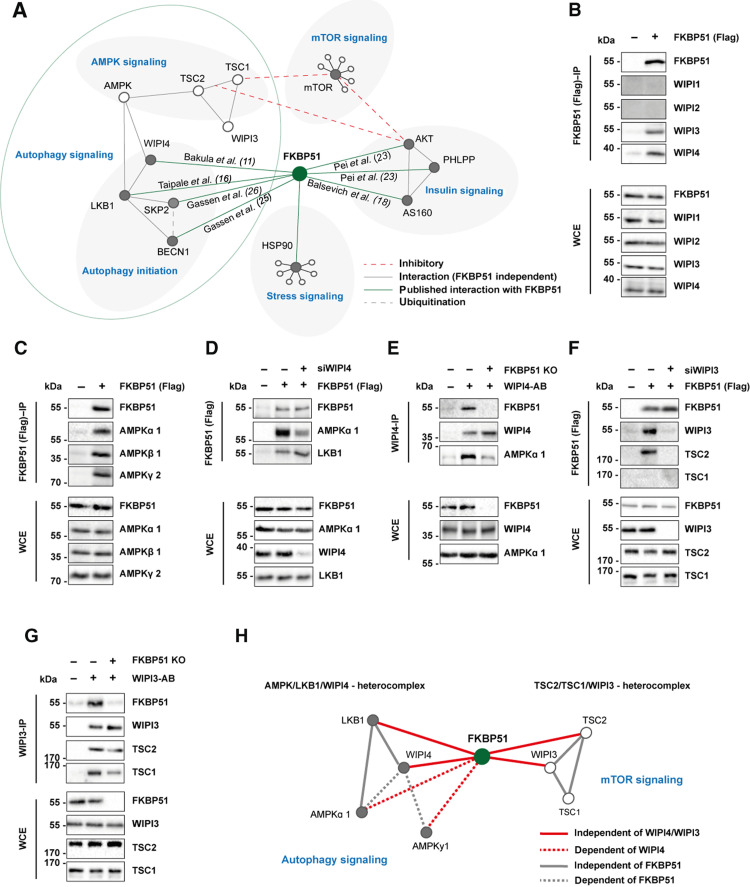
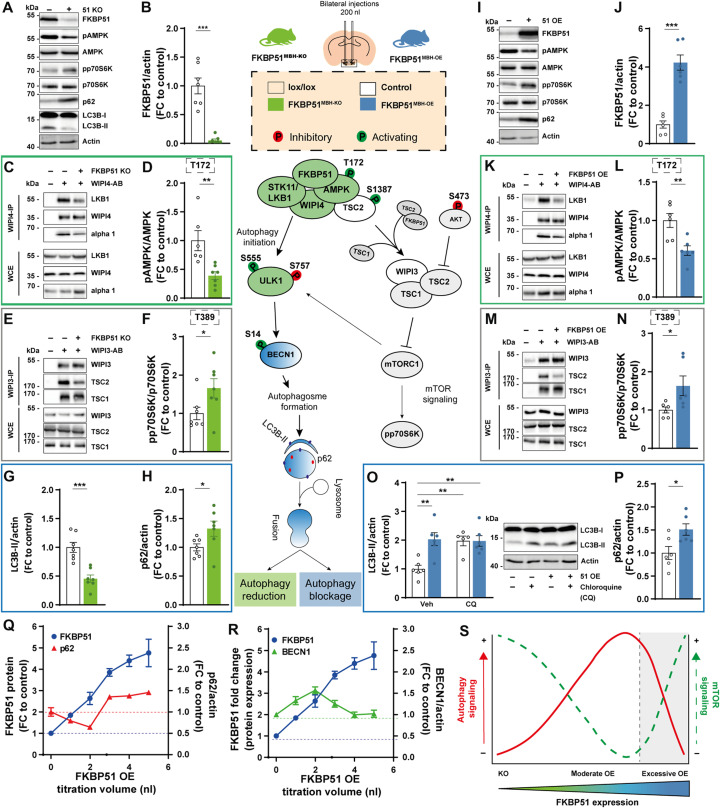
Fig. 3. FKBP51 associates with AMPK, TSC2, and WIPI3 and WIPI4 to regulate autophagy and mTOR signaling.
(A) Published protein-protein interactions of FKBP51. (B) FKBP51 associates with WIPI3 and WIPI4, but not with WIPI1 and WIPI2. WCE, whole-cell extract; IP, immunoprecipitation. (C) Interaction of FKBP51 with AMPK subunits. (D) Interaction of FKBP51 with LKB1 and AMPKα in WIPI4 KD cells. (E) Interaction of WIPI4 with AMPKα in FKBP51-lacking cells. (F) FKBP51 interacts with TSC2 in dependency of WIPI3. (G) WIPI3 interacts with TSC2 and TSC1 in the presence or absence of FKBP51. (H) Identified associations of FKBP51 in the regulation of autophagy and mTOR signaling and the proposed model of interaction. FKBP51 recruits LKB1 to the AMPK-WIPI4 complex and thereby facilitates AMPK activation. Furthermore, FKBP51 scaffolds TSC2-WIPI3 binding to alter mTOR signaling. AB, antibody.
FKBP51 associates with LKB1/AMPK/WIPI4 and TSC2/WIPI3 heteroprotein complexes to regulate autophagy and mTOR signaling
A recent study by Bakula and colleagues (11) investigated the role of the four WIPI proteins on autophagy, and the WIPI protein interactome revealed an association of WIPI4 with FKBP51, AMPKα1, and AMPKγ2 (11). WIPI proteins are essential scaffolding proteins that function as central molecular hubs to link key regulatory elements of autophagy with proteins that are sensitive to main metabolic cascades such as amino acid and glucose metabolism (41). On the basis of our FKBP51 starvation and OE experiments, we hypothesized that FKBP51 might scaffold WIPI4 and AMPK to induce autophagy initiation and therefore performed co-immunoprecipitation (co-IP) studies using N2a cells, with FKBP51-Flag OE.
These experiments confirmed that FKBP51 associates with WIPI4 but not with WIPI1 and WIPI2 (Fig. 3B), as suggested by Bakula and colleagues (11). Intriguingly, our experiments revealed a previously unidentified association of FKBP51 with WIPI3 (Fig. 3B). Next, we assessed the association of FKBP51 with various isoforms of AMPK and validated the expected interactions of FKBP51 with AMPKα1 and AMPKγ2 and further revealed a interaction with AMPKβ1 (Fig. 3C). We validated the previously unknown interactions of FKBP51 with WIPI3, WIPI4, AMPKα, and LKB1 in mouse WT N2a neuroblastoma cells performing IPs and co-IPs from endogenous proteins (fig. S5A).
To investigate the functional relevance of WIPI4 in the association of FKBP51 with AMPK, we generated WIPI4 knockdown (KD) N2a cells using small interfering RNA (siRNA) cotransfected with FKBP51-Flag (fig. S5B). We observed that FKBP51 binding to AMPK was reduced in cells lacking WIPI4 (Fig. 3D and fig. S5C). Our precipitation studies further demonstrated that FKBP51 interacts with LKB1, which confirmed the finding of a previous interactomics-based screening experiment by Taipale and colleagues (16) and further revealed that this interaction is independent of WIPI4 (Fig. 3D and fig. S5, D and A). Furthermore, we could demonstrate that WIPI4 deletion blocked the phosphorylation effect of FKBP51 OE on pAMPK at T172 (fig. S5E), suggesting that FKBP51 binds to AMPK in dependency of WIPI4. In addition, studies in FKBP51 KO cells revealed that the WIPI4 interaction to AMPK depends on the presence of FKBP51 (Fig. 3E), indicating a subordinate role of FKBP51 as a molecular bridge that links the autophagy-relevant WIPI network to AMPK signaling.
Last, we assessed the functional relevance of WIPI3-FKBP51 binding. Performing co-IP studies in N2a cells in the presence or absence of WIPI3 (fig. S5F), we observed that FKBP51 interacts with TSC2 but not TSC1 (Fig. 3F), which are upstream master regulators of mTOR (42). Further, we confirmed the association of FKBP51 with TSC2, which depends on the presence of WIPI3 (Fig. 3F), whereas the interaction of WIPI3 to TSC2 and TSC1 is independent of FKBP51 (Fig. 3G).
Collectively, we demonstrate that FKBP51 regulates autophagy in concert with the WIPI protein family, specifically WIPI3 and WIPI4 via interactions with the mTOR and AMPK cascades (Fig. 3H). The combined interpretation of our molecular and metabolomic analyses led us to hypothesize that FKBP51 regulates autophagy and consequently whole-body metabolism in vivo, especially after a metabolic challenge.
MBH FKBP51 is a regulator of body weight and food intake
We and others have previously shown that FKBP51 acts in a tissue-specific manner in soleus muscle (SM), epididymal WAT (eWAT), and the hypothalamus to regulate metabolism (18, 19, 22, 30). However, the interconnected response of autophagy and FKBP51 to a metabolic stressor, such as HFD, remains elusive and encouraged us to investigate the possible relationship between FKBP51 expression and autophagic flux (the protein turnover through catabolic autophagy).
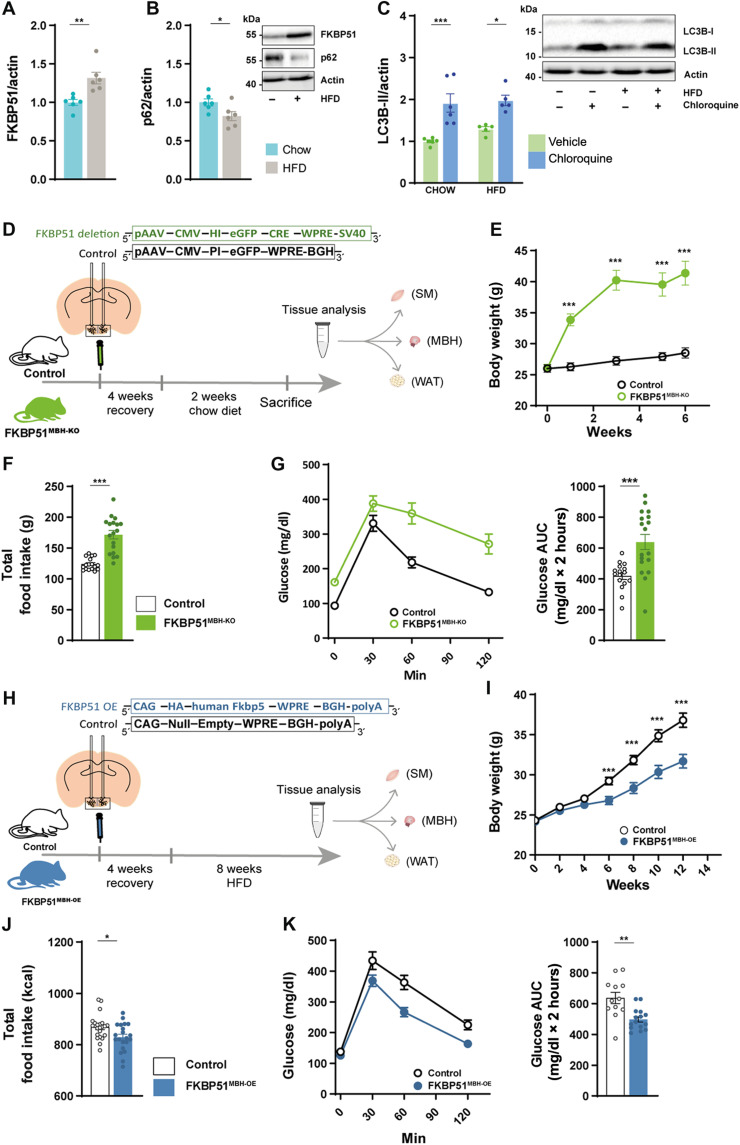
Our analysis revealed that C57BL/6N mice showed significantly increased FKBP51 protein levels (Fig. 4A) and diminished accumulation of the autophagy substrate p62 in the MBH upon 10 weeks of HFD (58% kcal from fat) (Fig. 4B). HFD per se did not affect the lipidation of the autophagosome-spiking protein light chain 3 (LC3B-I) to LC3B-II (Fig. 4C), which binds the autophagosome and is a reliable marker to analyze autophagic flux (35). However, levels of LC3B-II were similarly increased in both conditions after treatment with chloroquine (50 mg/kg), an inhibitor of lysosomal acidification and autophagosome-lysosome fusion that, in turn, blocks degradation of autophagosome cargo (35). In line with the reduced levels of p62, these results indicate an active central autophagic flux after HFD.
Fig. 4. MBH FKBP51 regulates body weight gain, food intake and glucose metabolism.
(A) Ten weeks of HFD increased hypothalamic FKBP51 in the MBH [n (chow) = 6 versus n (HFD) = 6]. (B) Effects of HFD on the accumulation of p62. (C) Treatment with chloroquine (50 mg/kg) increased LC3B-II level under chow and HFD conditions. (D) FKBP51lox/lox animals were injected with 200 nl of Cre-expressing virus and fed a chow diet for 6 weeks. (E) FKBP51MBH-KO showed significant body weight increase after virus injection on a regular chow diet. (F) FKBP51MBH-KO animals showed increased food intake and (G) enhanced glucose intolerance. AUC, area under the curve. (H) For FKBP51 overexpression, animals were injected with an AAV virus into the MBH. (I) FKBP51MBH-OE animals showed reduced body weight gain on an HFD diet compared to their control animals (J) FKBP51MBH-OE animals showed reduced food intake. (K) FKBP51MBH-OE animals showed improve glucose tolerance under HFD conditions. For (A), (B), (F), (G), (J), and (K), an unpaired Student’s t test was performed. For (C), a two-way ANOVA was performed, followed by a Tukey’s multiple comparison test. For (E) and (I), a repeated measurements ANOVA was performed. ± SEM; *P < 0.05, **P < 0.01, and ***P < 0.001.
In peripheral eWAT and SM, however, chloroquine treatment did increase LC3B-II levels only in chow-fed animals (fig. S6, A and B), implying reduced or even blocked autophagy signaling under HFD conditions. This hypothesis is supported by an increased accumulation of p62 in these peripheral tissues with a significant increase in SM (fig. S6C). FKBP51 protein levels were unaffected in eWAT and slightly but not significantly decreased in SM (fig. S6D). These data suggested a possible interconnected role of FKBP51 and autophagy flux, particularly in the MBH. We, therefore, decided to further test the effects of FKBP51 on autophagy signaling and its relevance for whole-body metabolism in vivo by manipulating FKBP51 in the MBH.
First, we injected a Cre-expressing virus into the MBH of FKBP51lox/lox animals (Fig. 4D) to evaluate the effects of central FKBP51 deletion. FKBP51MBH-KO animals showed a massive increase in body weight 6 weeks after surgery, despite their regular chow diet (Fig. 4E). The bodyweight increase was accompanied by increased food intake and decreased glucose tolerance (Fig. 4, F and G). These findings were unexpected, considering the lean phenotype of full-body FKBP51-deficient mice after prolonged exposure to an HFD (18, 19) and further highlight the tissue specificity of FKBP51.
Next, we injected an adeno-associated virus (AAV)–mediated FKBP51 OE virus into the MBH of C57/Bl6 mice (FKBP51MBH-OE, Fig. 4H). FKBP51MBH-OE animals showed no substantial differences in body weight gain within the first 4 weeks after surgery. Therefore, we challenged FKBP51MBH-OE animals with a metabolic stressor by feeding them an HFD for 8 weeks. Animals overexpressing FKBP51 displayed significantly reduced body weight gain compared to the control group (Fig. 4I), paralleled by a reduction in food intake (Fig. 4J). In a second cohort of FKBP51MBH-OE animals [with an identical body weight phenotype (fig. S6E)], we investigated whether glucose metabolism was altered and observed improved glucose tolerance and insulin sensitivity compared to the control group under HFD but not under normal chow diet (Fig. 4K and fig. S6, F to H). Together, these experiments reveal an essential role of MBH FKBP51 in central coping mechanisms with an obesogenic stressor and position MBH FKBP51 as a key regulator of whole-body metabolism.
MBH FKBP51 fine-tunes autophagy signaling in an inversed U-shaped manner
Given the opposing phenotypes of FKBP51MBH-KO and FKBP51MBH-OE animals and the discovered regulatory function of FKBP51 in the metabolic control of autophagy, we were interested in the underlying regulation of autophagy signaling. In our KO experiment, viral injection resulted in a high-deletion rate within the MBH of FKBP51MBH-KO animals (Fig. 5, A and B). According to our hypothesis, we observed a reduced binding of LKB1 and AMPK to WIPI4 (Fig. 5C and see fig. S7A for quantification), which was accompanied by a reduction in the phosphorylation of AMPK at T172, causing less active AMPK (Fig. 5D). Downstream of AMPK, we monitored diminished phosphorylation of ULK1 at S555, BECN1 at S91/S94, and TSC2 at S1387 (fig. S7, B to D). Parallel to the effects of AMPK downstream proteins, we detected reduced levels of TSC2 binding to WIPI3 (Fig. 5E and see fig. S7E for quantification). Furthermore, we observed increased levels of phosphorylated AKT at S473 and phosphorylated ULK1 at S757 (fig. S7, F and G), indicating increased AKT/mTOR signaling. The increased mTOR activity could be validated by increased levels of pp70S6K (Fig. 5F). Last, loss of FKBP51 resulted in decreased levels of LC3B-II and the accumulation of p62 (Fig. 5, G and H). Together, these data suggest that FKBP51 deletion reduced autophagy signaling in the MBH via the reduction of AMPK activity and an increased mTOR signaling, which is in line with our in vitro data.
Fig. 5. MBH FKBP51 regulates autophagy in an inversed U-shaped manner.
FKBP51 deletion is depicted in green, and FKBP51 overexpression is depicted in blue. (A) Representative blots of autophagy and mTOR markers in FKBP51MBH-KO mice. (B) Quantification of FKBP51 deletion. (C) FKBP51 deletion reduced LKB1 and AMPK binding to WIPI4 as well as (D) AMPK phosphorylation at T172. (E) TSC2-WIPI3 binding was decreased in FKBP51MBH-KO animals. (F) Quantification of mTOR substrate pp70S6K (T389). (G) LC3B-II and (H) p62 levels in the MBH. (I) Representative blots of autophagy and mTOR marker in FKBP51MBH-OE mice. (J) Quantification of viral FKBP51 overexpression. (K) FKBP51 overexpression reduced LKB1 and AMPK binding to WIPI4. (L) Quantification of AMPK phosphorylation at T172. (M) TSC2-WIPI3 binding was decreased. (N) Quantification of pp70S6K phosphorylation at T389. (O) To assess autophagic flux FKBP51MBH-OE, animals were treated with chloroquine (50 mg/kg), and LC3B-II levels were analyzed 4 hours after treatment. (P) FKBP51 overexpression blocked autophagic flux and resulted in an accumulation of p62. (Q and R) Quantification of FKBP51, p62, and BECN1, while titrating AAV-HA-FKBP51 virus into mouse neuroblastoma cells. (S) MBH FKBP51 regulates autophagy and mTOR signaling in a dose-dependent manner. All data are shown as ±SEM. Data are shown as the relative protein expression compared to control; for (A) to (N), an unpaired Student’s t test was performed. *P < 0.05, **P < 0.01, and ***P < 0.001.
Animals overexpressing FKBP51 in the MBH showed an excessive up-regulation of FKBP51 (Fig. 5, I and J). Co-IP studies indicated that following the excessive overexpression (OE) of FKBP51, AMPK binding to WIPI4 was decreased. Furthermore, LKB1 levels were strongly reduced and binding of LKB1 to WIPI4 vanished (Fig. 5K and fig. S7A). Consequently, phosphorylation of AMPK at T172 was significantly reduced (Fig. 5L). Downstream of AMPK, we observed a decrease in phosphorylation of ULK1 at S555 and no changes of phosphorylated BECN1 levels (fig. S7, B and C). Phosphorylation of TSC2 at S1387 and the binding of TSC2 to WIPI3 were significantly decreased (Fig. 5M and fig. S7, D and E). Phosphorylation of AKT at S473 was unchanged (fig. S7F). On the other hand, FKBP51MBH-OE animals showed increased mTOR signaling, indicated by increased phosphorylation of ULK1 at S757 and elevated levels of pp70S6K (Fig. 5N and fig. S7G). Last, FKBP51 OE animals showed increased levels of LC3B-II. However, treatment with chloroquine (50 mg/kg) did not further increase the LC3B-II levels in FKBP51MBH-OE animals (Fig. 5O), indicating that the fusion of autophagosomes with lysosomes is impaired and central autophagic flux is blocked. This hypothesis is supported by the fact that FKBP51 OE resulted in the accumulation of the autophagy substrate p62 (Fig. 5P). These data are in contrast to our previously observed findings and imply that FKBP51MBH-OE animals, despite their highly elevated FKBP51 levels, have blocked autophagy signaling in the MBH.
The observation that viral overexpression of FKBP51 in the MBH resulted in a massive overexpressing of FKBP51 led us to hypothesize that the level of FKBP51 expression directly correlates with the degree of autophagy signaling. Therefore, we gradually increased FKBP51 levels in N2a cells by the titration of AAV–hemagglutinin tag (HA)–FKBP51. Following a moderate increase of FKBP51, we observed a decrease in the accumulation of p62 and an increase in BECN1, supporting the activating role of FKBP51. In parallel, we could observe a slight decrease in p70S6K phosphorylation. However, upon a stimulus threshold (at approximately three- to fourfold FKBP51), we observed an inhibitory effect on autophagy with decreased protein level of BECN1 and an increased phosphorylation of p70S6K and enhanced accumulation of p62 (Fig. 5, Q and R, and fig. S7H).
Our pathway analysis demonstrates that deletion of FKBP51 reduces autophagy signaling, and excessive levels of FKBP51 protein results in a total block of autophagy, causing a substantial shift from autophagy to mTOR signaling. In conclusion, we suggest that FKBP51 dose dependently regulates autophagy signaling in an inversed U-shaped manner (Fig. 5S).
MBH FKBP51 alters sympathetic outflow and thereby regulates autophagy signaling in the periphery
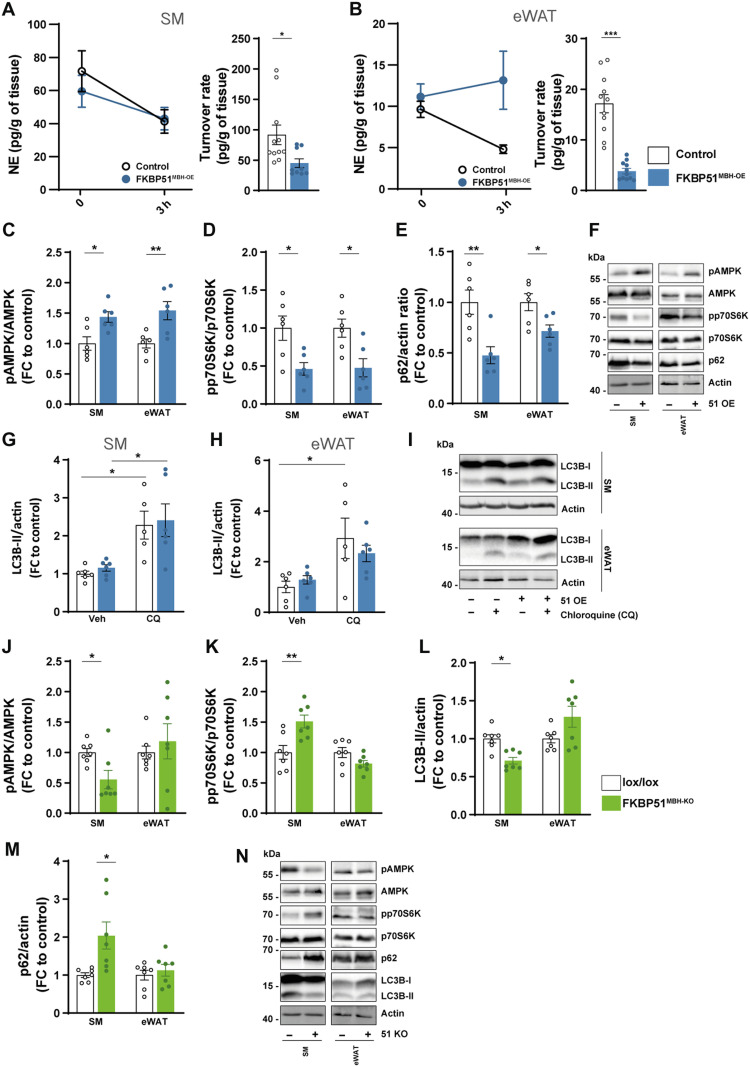
The MBH is an established regulatory center for sympathetic outflow to peripheral tissues (43). Consequently, we were interested whether the sympathetic tone of the brain into peripheral tissues was affected in FKBP51MBH-OE animals. To do so, we treated FKBP51MBH-OE mice with a single dose of the norepinephrine (NE) synthesis inhibitor α-methyl-p-tyrosine (α-MPT) to block NE synthesis in the periphery, thereby enabling the assessment of the catecholamine turnover rate (cTR) (44, 45). MBH FKBP51 OE led to a reduction in the cTR in muscle and eWAT (Fig. 6, A and B). Further, inguinal WAT (iWAT) of FKBP51MBH-OE animals showed significant differences in initial NE levels, but not in cTR (fig. S8A). We also observed a mildly but not significantly decreased cTR in brown adipose tissue (BAT) (fig. S8B), whereas no effects were detected in the pancreas or heart tissue (fig. S8, C and D). Together, these data demonstrate that MBH FKBP51 OE dampens the sympathetic outflow especially to muscle and fat tissue and encouraged us to investigate changes in autophagy signaling in both tissues.
Fig. 6. MBH FKBP51 affects sympathetic outflow and peripheral autophagy signaling.
FKBP51 overexpression is depicted in blue, and FKBP51 deletion is depicted in green. (A and B) Representative decrease in tissue NE content after α-MPT injection (left) and turnover rate (right) were determined on SM and eWAT (see fig. S8 for pancreas, heart, iWAT, and BAT tissues). Quantification of (C) pAMPK (T172) and (D) pp70S6K (T389), and (E) p62 level in the SM and eWAT. (F) Representative blots. (G to H) FKBP51 overexpression increased autophagic flux and in SM and eWAT. (I) Representative blots of chloroquine the experiment. Quantification of (J) pAMPK (T172), (K) pp70S6K (T389), (L) LC3B-II, and (M) p62 levels in SM and eWAT in animals lacking FKBP51 in the MBH. (N) Representative blots of FKBP51MBH-KO protein analysis. All data are shown as ±SEM. Protein data are shown as the relative protein expression compared to control. A two-way ANOVA was performed, followed by a Tukey’s multiple comparison test in (F) and (G). For (A) to (E) and (I) to (L), an unpaired Student’s t test was performed. *P < 0.05, **P < 0.01, and ***P < 0.001.
In FKBP51MBH-OE animals, we observed increased levels of FKBP51 in SM and eWAT (fig. S8E), which resulted in increased AMPK activity (Fig. 6C) by enhanced binding of AMPK and LKB1 to WIPI4 (fig. S8, F and G). Downstream of AMPK, we observed increased phosphorylation of ULK1, BECN1, and TSC2, indicating enhanced autophagy initiation in the periphery (fig. S8, H to J). We again assessed mTOR signaling and observed increased binding of TSC2 to WIPI3 (fig. S8K) and a strong reduction in the phosphorylation of AKT at S473 and ULK1 at S757 (fig. S8, L and M), which resulted in reduced levels of pp70S6K (T389) (Fig. 6D). Furthermore, we monitored reduced levels of p62 (Fig. 6, E and F). To verify the increase in autophagy signaling, we analyzed LC3B-II levels before and after chloroquine treatment. Here, we detected a true increase in LC3B-II levels after chloroquine treatment (Fig. 6, G to I). These data imply an increase in autophagy flux in the periphery of FKBP51MBH-OE animals. This is in line with our hypothesis that moderately elevated levels of FKBP51 increase autophagy signaling and suggest that the balance between active mTOR signaling in the MBH and active autophagy signaling in the periphery is one driving factor of the lean phenotype of the FKBP51MBH-OE animals.
In FKBP51MBH-KO mice, we observed an opposing phenotype with reduced autophagy signaling in SM, whereas autophagy signaling in eWAT was unaltered. In both tissues, we did not observe significant changes in FKBP51 protein level (fig. S8N). However, we could detect less phosphorylation of AMPK at T172 (Fig. 6J) and reduced binding of AMPK/LKB1 to WIPI4 (fig. S8, O and P). These findings were accompanied by reduced levels of ULK1, BECN1, and TSC2 (fig. S8, Q to S). Furthermore, we monitored increased levels of pp70S6K (T389), pULK1 (S757), and pAKT (S473), suggesting increased mTOR signaling (Fig. 6K and fig. S8, U and V). Last, LC3B-II levels were significantly reduced (Fig. 6L) in combination with elevated levels of p62 in SM (Fig. 6, M and N), which is indicative of reduced autophagy signaling solely in this peripheral tissue. We could not detect any differences in autophagy signaling in other peripheral tissues, such as the liver (fig. S9, A and B). In summary, we suggest that the combined reduction of autophagy in the MBH and peripheral tissues, such as muscle and adipose tissue, is driving the observed body weight phenotype in FKBP51MBH-KO mice.
DISCUSSION
In the current study, we examined the role of stress-activated chaperone FKBP51 as a molecular master switch linking autophagy and whole-body metabolism. We here present that FKBP51 actively modulates the response of the AMPK-mTOR network to an HFD by scaffolding autophagy-upstream AMPK/LKB1/WIPI4 and TSC2/WIPI3 heteroprotein complexes. We identify a tissue-specific function of FKBP51 by providing in vivo evidence that hypothalamic FKBP51 acts as a dose-specific mediator of whole-body metabolism.
Metabolomic profiling of neuronal-like cells lacking FKBP51 revealed a substantial increase for several metabolites and amino acids and suggests a role of FKBP51 in BCAA metabolism. BCAAs are important regulators of neurotransmitters and protein synthesis as well as food intake (46, 47). The increase of multiple BCAAs have been associated with obesity and insulin resistance (48, 49). In our in vitro metabolomic profiling analysis, isoleucine, leucine, valine, and tyrosine were strongly elevated in FKBP51 KO cells under normal and high glucose concentrations, which is indicative of constantly active mTOR signaling (50). Furthermore, it has been shown that excess leucine can reduce abdominal fat loss, whereas leucine deprivation promotes fat loss via cyclic AMP response element-binding protein signaling and increased expression of CRH (corticotropin-releasing hormone) in the hypothalamus. This effect is conveyed by the activation of the sympathetic nervous system (51). Leucine is an important regulator of mTOR and negatively affects the biogenesis of autophagosomes through its metabolite acetyl coenzyme A, which thereby enhances acetylation of the regulatory-associated protein of MTOR (RPTOR) via acetyltransferase EP300 in neurons and other cell types. This cascade of events ultimately leads to autophagy inhibition and mTOR activation (52, 53). At the same time, the increased levels of polyamines, observed in FKBP51 KO cells, are in contrast to autophagy inhibition. In particular, spermidine was shown to be capable of autophagy induction via inhibition of EP300 (54). Nevertheless, cell type–specific effects have to be taken into account as studies suggest an increased expression of EP300 in response to spermidine supplementation in aged and osteoarthritic chondrocytes (55).
To gain further insight into the underlying mechanisms, we built on already existing knowledge about FKBP51 regarding its regulatory role on single autophagy-related proteins [like BECN1, WIPIs, and SKP2 (11, 16, 26)], which further positions FKBP51 as a major upstream regulator of autophagy. AMPK is activated by the phosphorylation of T172, which is regulated by LKB1 (56), and increased LKB1/AMPK signaling activates the TSC1/TSC2 complex, which, in turn, inhibits mTOR activity (38). Recently, Bakula and colleagues (11) showed that the WIPI protein family members WIPI3 and WIPI4 are essential scaffolders of the LKB1/AMPK/TSC1/2 signaling network thereby regulating autophagy and mTOR signaling. Here, we extended this knowledge by revealing that FKBP51 recruits LKB1 to the WIPI4-AMPK regulatory platform to induce AMPK phosphorylation at T172, which further increases autophagy initiation by direct phosphorylation of ULK1 at S555 (10). On the other hand, FKBP51 associates with the TSC2/WIPI3 heterocomplex to coregulate mTOR signaling and thus position FKBP51 as a main regulatory switch between autophagy initiation and mTOR signaling. Our experiments in vivo and in vitro led us to propose a model in which physiological levels of FKBP51 are essential for normal cellular autophagy-mediated homeostasis. Hereby, the absence of FKBP51 reduces autophagy signaling and capacity in contrast to excessive, nonphysiological levels of FKBP51, which block autophagy in favor of mTOR signaling. The relative amounts of FKBP51 complexed with autophagy regulators within a cell govern the threshold for a transition from cell homeostasis to impaired autophagy in an inversed U-shaped manner.
FKBP51 has been shown to act tissue-specific to control adipocyte differentiation, browning (19, 57), and glucose metabolism (18). Most studies investigated global FKBP51 KO mice and observed a lean phenotype after an HFD regimen. These observations would imply that hypothalamic FKBP51 OE increases body weight gain, whereas deletion reduces body weight. We here observed that the acute FKBP51 manipulation in the MBH acts in an opposing manner with a central deletion leading to obesity and an overexpression to a lean phenotype, emphasizing its tissue specificity. These data suggest that hypothalamic FKBP51 regulates body weight and food intake in a U-shaped manner, which is supported by the fact that HFD increases FKBP51 expression and that a modest increase of hypothalamic FKBP51 induces body weight gain (30).
Several studies have already addressed the functional role of hypothalamic autophagy in the regulation of whole-body metabolism. Mice challenged with a chronic HFD showed impaired autophagy in the arcuate nucleus, and a deletion of ATG7 in the MBH resulted in hyperphagia and increased body weight (58). The MBH, however, is a complex brain region with multiple different nuclei, which have various opposing roles in the control of whole-body metabolism (1, 59). Here, we targeted FKBP51 nonselectively in various nuclei within the MBH, which is a limitation of the study. However, it also raises the question, which nuclei or neuronal subpopulations are the driving force behind our observed phenotypes. Previous studies have shown that specific deletion of autophagy in POMC neurons leads to hyperphagia and obesity (12). These findings are in line with our data of the FKBP51MBH-KO animals, which develop obesity and hyperphagia on a regular chow diet in combination with decreased activity of central autophagy. Intriguingly, animals overexpressing FKBP51 in the MBH have a reduced body weight progression, increased glucose tolerance, and insulin sensitivity under an HFD regimen despite the hypothalamic autophagy blockage. The deletion of autophagy in AgRP neurons resulted in decreased body weight and food intake in response to fasting (13). The current study, however, cannot fully address whether the observed effects on obesity and the sympathetic effect output are purely driven by FKBP51 manipulation and whether autophagy is required for this effect. Future studies should emphasize the specific neuronal action of FKBP51 on autophagy to regulate body weight progression and food intake and investigate whether a blockade of autophagy can indeed counteract the effects of FKBP51 overexpression and vice versa.
Last, we suggest that the altered balance between hypothalamic and peripheral autophagy–mTOR signaling is a major contributor to the observed phenotype of FKBP51MBH-OE and FKBP51MBH-KO animals. FKBP51MBH-KO animals showed decreased autophagy and increased mTOR signaling in the periphery (eWAT and SM), whereas peripheral autophagy signaling in FKBP51MBH-OE animals displayed the opposite phenotype. The importance of central-peripheral mTOR and autophagy signaling has already been studied intensely. For instance, peripheral mTOR activity was shown to be involved in the pathogenesis of obesity and is enhanced in muscle and adipose tissue in obese animals (60, 61), whereas the activation of central mTOR can reduce food intake and body weight gain (62). In particular, the mTOR substrate p70S6K was shown to regulate body weight by mediating the sensitivity of leptin to AMPK via PI3K/AKT1/mTOR pathway (63, 64). Here, we extend this finding by the fact that the combination of peripheral and central p70S6K activity is an important contributor to the development of obesity. One has to keep in mind, although, that FKBP51 is highly dynamically regulated by stressful situations and acts in concert with other chaperone proteins in a cell type– and tissue-specific manner. This may also explain apparent minor inconsistencies in our data and warrants further investigation.
In conclusion, this study provides a conceptual framework for the regulatory function of the stress-responsive co-chaperone FKBP51 on autophagy signaling and establishes a physiological role of MBH FKBP51 in the regulation of food intake and body weight regulation. We further suggest that FKBP51 is a crucial sensor linking signaling pathways controlling the stress response, autophagy, and metabolism. The ability of FKBP51 to regulate autophagy and energy homeostasis might therefore open new promising treatment avenues for metabolic disorders, such as obesity and type 2 diabetes.
MATERIALS AND METHODS
Antibodies
The following antibodies were used: goat polyclonal anti-actin (I-19) (sc-1616, Santa Cruz Biotechnology), rabbit polyclonal anti-FKBP51 (A301-430A, Bethyl Laboratories), rabbit monoclonal anti-FKBP5 (D5G2, #12210, Cell Signaling Technology), rabbit monoclonal anti-LKB1 (D60C5, #3047, Cell Signaling Technology), rabbit polyclonal anti-pAMPKαT172 (#2531, Cell Signaling Technology), rabbit polyclonal anti-pAMPKα (#2532, Cell Signaling Technology), rabbit polyclonal anti-SKP2 (L70, #4313, Cell Signaling Technology), rabbit anti-pSKP2S72 (was a gift from Cell Signaling Technology), rabbit polyclonal anti-AKT (#9272, Cell Signaling Technology), rabbit monoclonal anti-pAKTS473 (D9E, #4060, Cell Signaling Technology), rabbit polyclonal anti-p62 (#5114, Cell Signaling Technology), rabbit monoclonal anti-LC3B (D11, #3868, Cell Signaling Technology), rabbit polyclonal anti-pULK1S757 (#6888, Cell Signaling Technology), rabbit monoclonal anti-pULK1S555 (D1H4, #5869, Cell Signaling Technology), rabbit monoclonal anti-ULK1 (D8H5, #8054, Cell Signaling Technology), anti-pBECN1S93/S96 (in mouse S91/S94) (#12476, Cell Signaling Technology), rabbit polyclonal anti-pBECN1S15 (#84966, Cell Signaling Technology), rabbit polyclonal anti-BECN1 (#3738, Cell Signaling Technology), rabbit polyclonal anti-TSC2 (#3612, Cell Signaling Technology), rabbit polyclonal anti-pTSC2S1387 (#5584, Cell Signaling Technology), rabbit monoclonal anti-pATG16L1S278 (EPR19016, ab195242, Abcam), rabbit polyclonal anti-WIPI4 (WDR45) (19194-1-AP, Proteintech), mouse monoclonal anti-WIPI4 (G12, sc-398272, Santa Cruz Biotechnology), rabbit polyclonal anti-WIPI3 (WDR45L) (SAB2102704, Sigma-Aldrich), mouse monoclonal anti-WIPI3 (B-7, sc-514194, Santa Cruz Biotechnology), rabbit polyclonal anti-WIPI2 (#8567, Cell Signaling Technology), rabbit polyclonal anti-WIPI1 (HPA007493, Sigma-Aldrich), rabbit polyclonal anti-AMPKα1 (#2795, Cell Signaling Technology), rabbit polyclonal anti-AMPKγ2 (#2536, Cell Signaling Technology), rabbit polyclonal anti-AMPKα2 (#2757, Cell Signaling Technology), rabbit monoclonal anti-AMPKβ1 (71C10, #4178, Cell Signaling Technology), rabbit polyclonal anti-AMPKγ1 (#4187, Cell Signaling Technology), rabbit polyclonal anti-AMPKβ2 (#4188, Cell Signaling Technology), rabbit polyclonal anti-AMPKγ3 (#2550, Cell Signaling Technology), rabbit monoclonal anti-TSC1 (D43E2, #6935, Cell Signaling Technology), rabbit polyclonal anti-Flag (600-401-383, Rockland Inc.), rabbit polyclonal anti-hypusine (ABS1046, Merck Millipore), rabbit monoclonal anti-eIF5A (D8L8Q, #20765, Cell Signaling Technology), and rabbit polyclonal anti-TFEB (ab245350, Abcam).
Animals and animal housing
All experiments and protocols were approved by the committee for the Care and Use of Laboratory Animals of the Government of Upper Bavaria and were performed in accordance with the European Communities’ Council Directive 2010/63/EU. All animals were kept singly housed in individually ventilated cages (IVCs) (30 cm by 16 cm by 16 cm; 501 cm2) with ad libitum access to water and food and constant environmental conditions (12-hour light/12-hour dark cycle, 23° ± 2°C, and humidity of 55%) during all times. All IVCs had sufficient bedding and nesting material as well as a wooden tunnel for environmental enrichment. All animals were fed with a standard research chow diet (Altromin 1318, Altromin GmbH, Germany) or an HFD (58% kcal from fat; D12331, Research Diets, New Brunswick, NJ, USA). For all experiments, male C57BL/6N or male Fkbp5lox/lox mice [described in (65)] aged between 2 and 5 months were used.
Viral overexpression and knockdown of FKBP51
For overexpression of FKBP51, we injected an AAV vector containing a CAG-HA–tagged FKBP51-WPRE-BGH-polyA expression cassette (containing the coding sequence of human FKBP51 National Center for Biotechnology Information CCDS ID CCDS4808.1) in C57BL/6N mice. The same vector construct without expression of FKBP51 (CAG-Null/Empty-WPRE-BGH-polyA) was used as a control. Virus production, amplification, and purification were performed by GeneDetect. A viral vector containing a Cre-expressing cassette (pAAV-CMV-HI-eGFP-Cre-WPRE-SV40, #105545, Addgene) was used to induce FKBP51 deletion in FKBP51lox/lox mice. Control animals were injected with a control virus (pAAV-CMV-PI-eGFP-WPRE-bGH, #105530, Addgene). For both experiments, stereotactic injections were performed as described previously (66). Briefly, mice were anesthetized with isoflurane prior surgery, and 0.2 μl of the abovementioned viruses (titers: 1.6 × 1012–13 genomic particles/ml) was bilaterally injected in the MBH at 0.05 μl/min by glass capillaries with a tip resistance of 2 to 4 megohm in a stereotactic apparatus. The following coordinates were used: −1.5 mm anterior to bregma, 0.4 mm lateral from midline, and 5.6 mm below the surface of the skull, targeting the MBH. After surgery, mice were treated for 3 days with Metacam via intraperitoneal injections and were housed for 3 to 4 weeks for total recovery before the actual experiments. Successful overexpression and KD of FKBP51 were verified by Western blot.
Autophagic flux
We investigated the autophagic flux by the injection of chloroquine (50 mg/kg), an inhibitor of lysosomal acidification and autophagosome-lysosomal fusion that blocks degradation of autophagosome cargo (35). We injected C57BL/6N mice in the morning with chloroquine (50 mg/kg) or saline as control. Multiple tissues were removed and shock-frozen 4 hours after injection and stored at −80°C until protein analysis of LC3B-II normalized to glyceraldehyde-3-phosphate dehydrogenase or actin. Lipidation of LC3B in protein homogenates obtained from animals treated with chloroquine (fusion block) was compared to LC3B lipidation of animals treated with vehicle.
Sample collection
On the day of euthanization, animals were deeply anesthetized with isoflurane and euthanized by decapitation. Trunk blood was collected in labeled 1.5-ml EDTA-coated microcentrifuge tubes (Sarstedt) and kept on ice until centrifugation. After centrifugation (4°C, 8000 rpm for 1 min), the plasma was removed and transferred to new, labeled tubes and stored at −20°C until hormone quantification. For protein analysis, the MBH, skeletal muscle (SM), and WAT (eWAT) were collected and immediately shock-frozen and stored at −80°C until protein analysis.
cTR determination
Catecholamine turnover was measured on the basis of the decline in tissue NE content after the inhibition of catecholamine biosynthesis with α-methyl-dl-tyrosine (α-MPT) (200 mg/kg i.p. injection; Sigma-Aldrich, ST, Quentin, France), as described previously (44).
In the morning, bedding was changed, and C57BL/6N mice were food-deprived for 3 hours to insure postprandial state and injected with α-methyl-dl-tyrosine (α-MPT; a tyrosine hydroxylase inhibitor) to block catecholamine synthesis. Before (time = 0) and 3 hours after the injection (time = 3 hours), animals were euthanized, and the tissues were removed, flash-frozen in liquid nitrogen, and stored at −80°C for monoamine and metabolite analysis.
Catecholamine content at time = 0 [NE (0)] was determined on a group of animals receiving a saline injection. Because the concentration of catecholamine in tissues declined exponentially, we could obtain the rate constant of NE efflux (expressed in h − 1). Comprehensive analysis of NE was carried out by reverse-phase liquid chromatography (LC) with electrochemical detection as described in (67). The values obtained were expressed as nanogram per milligram wet tissue and were logarithmically transformed for calculation of linearity of regression, SE of the regression coefficients, and significance of differences between regression coefficients.
Glucose tolerance and insulin tolerance
Alteration of glucose metabolism in FKBP51MBH-OE and FKBP51MBH-KO mice was investigated by a glucose (glucose tolerance test) and insulin tolerance (insulin tolerance test) test as described previously (18).
Hormone assessment
Corticosterone concentrations were determined by radioimmunoassay using a corticosterone double antibody 125I radioimmunoassay kit (sensitivity: 12.5 ng/ml; MP Biomedicals Inc.) and were used following the manufacturers’ instructions. Radioactivity of the pellet was measured with a gamma counter (Packard Cobra II Auto Gamma, PerkinElmer). Final corticosterone levels were derived from the standard curve.
Cell lines and transfection
N2a WT, N2a FKBP51 KO, SH-SY5Y WT, and FKBP51 KO (68) cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and 1× penicillin-streptomycin antibiotics at 37°C in a humidified atmosphere with 5% CO2. At 90% confluency, N2a cells were detached from the plate, and 2 × 106 cells were resuspended in 100 μl of transfection buffer [50 nM Hepes (pH 7.3), 90 mM NaCl, 5 mM KCl, and 0.15 mM CaCl2]. A total of 2.5 μg of plasmid DNA or 80 ng of siRNA (siWIPI3, EMU081491 or siWIPI4, EMU007321 or siControl, and SIC001, all Sigma-Aldrich) was used per transfection. Electroporation was performed using the Amaxa Nucleofector System 2b (program T-020). For OE experiments, N2a cells were transfected with FKBP51-Flag expression or TFEB-GFP reporter plasmid using Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s instructions.
Generation of FKBP51 KO N2a cells
N2a (Sigma-Aldrich) FKBP51 KO cell line was generated with the Alt-R CRISPR-Cas9 System from Integrated DNA Technologies (IDT) according to the manufacturer’s instructions. Briefly, RNA oligos [Alt-R CRISPR-Cas9 crispr RNA (crRNA) against murine FKBP51 and Alt-R CRISPR-Cas9 trans-activating crRNA (tracrRNA)] were mixed in nuclease-free duplex buffer (IDT) in equimolar concentrations yielding a final duplex of 1 μM and then heated at 95°C for 5 min and combined with 1 μM Alt-R S.p. HiFi Cas9 Nuclease V3 diluted in Opti-MEM (Thermo Fisher Scientific). Ribonucleoprotein (RNP) complexes were assembled at room temperature (RT) for 5 min and mixed with Lipofectamine RNAiMAX reagent (Thermo Fisher Scientific) and Opti-MEM (Thermo Fisher Scientific) and incubated for 20 min at RT to form transfection complexes. Subsequently, 40,000 N2a cells per well were reverse-transfected using complete culture media without antibiotics in a 96-well tissue culture plate with a final RNP concentration of 10 nM. After 48 hours (37°C, 5% CO2), single-cell clones were obtained by array dilution method, expanded, and analyzed by Western blotting. FKBP51 WT control cells were identified by immunoblotting after single-cell cloning procedures and, therefore, underwent the same transfection and isolation procedure as the FKBP51 KO cells. Predesigned Alt-R CIRSPR Cas9 guide RNAs (IDT) were used for KO generation (protospacer adjacent motif sequence in italics). Mm.Cas9.FKBP5.1.AA: CGATCCCAATCGGAATGTCGTGG.
Treatment of N2a cells
Treatments of N2a cell cultures included glutocorticoid receptor (GR) stimulation with dexamethasone (Sigma-Aldrich) ranging from 1 to 100 nM for 24 hours, HBSS (Thermo Fisher Scientific)–induced starvation for 4 hours, and inhibition of autophagosome-lysosome fusion by BafA1 (100 nM, 4 hours; Alfa Aesar).
Co-immunoprecipitation
IPs of endogenous proteins were performed from protein extracts (n = 3 to 4 per group) derived from N2a cells, SH-SY5Y WT or FKBP51 KO cells, SM, eWAT, and MBH. For co-IPs, 500 μg of lysate was incubated with 2 μg of the appropriate IP antibody [anti-Flag (FKBP51), anti-FKBP51, anti-WIPI4, and anti-WIPI3] at 4°C overnight. A total of 20 μl of rabbit immunoglobulin G–conjugated protein G Dynabeads (Invitrogen, 100-03D) were blocked with bovine serum albumin and subsequently added to the lysate-antibody mixture and allowed to incubate at 4°C for 3 hours to mediate binding between Dynabeads and the antibody-antigen complex of interest. Beads were then washed three times with ice-cold phosphate-buffered saline, and the protein antibody complexes were eluted with 60 μl of Laemmli loading buffer. Thereafter, the eluate was boiled for 5 min at 95°C. Then, 2 to 5 μl of each immunoprecipitate were separated by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and electro-transferred onto nitrocellulose membranes. For assessing protein complexes, immunoblotting against WIPI1-WIPI4, FKBP51, LKB1, AMPK, TSC1, and TSC2 was performed.
Western blot analysis
Protein extracts were obtained by lysing cells [in radioimmunoprecipitation assay buffer (150 mM NaCl, 1% IGEPAL CA-630, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mM tris (pH 8.0)] freshly supplemented with protease inhibitor (Merck Millipore, Darmstadt, Germany), benzonase (Merck Millipore), 5 mM dithiothreitol (Sigma-Aldrich, Munich, Germany), and phosphatase inhibitor cocktail (Roche, Penzberg, Germany). Proteins were separated by SDS- PAGE and electro-transferred onto nitrocellulose membranes. Blots were placed in tris-buffered saline supplemented with 0.05% Tween (Sigma-Aldrich) and 5% nonfat milk for 1 hour at RT and then incubated with primary antibody (diluted in tris-buffered saline/0.05% Tween) overnight at 4°C.
Subsequently, blots were washed and probed with the respective horseradish peroxidase or fluorophore-conjugated secondary antibody for 1 hour at RT. The immunoreactive bands were visualized either using an enhanced chemiluminesence detection reagent (Millipore, Billerica, MA, USA) or directly by excitation of the respective fluorophore. Determination of the band intensities was performed with Bio-Rad, ChemiDoc MP.
LC-MS analysis of amine-containing metabolites
The benzoyl chloride derivatization method was used for amino acid analysis (69). Briefly, the dried metabolite pellets were resuspended in 90 μl of the LC-MS grade water (Milli-Q 7000 equipped with an LC-Pak and a Millipak filter, Millipore). Then, 20 μl of the resuspended sample was mixed with 10 μl of 100 mM sodium carbonate (Sigma-Aldrich), followed by the addition of 10 μl of 2% benzoyl chloride (Sigma-Aldrich) in acetonitrile (Optima-Grade, Fisher Scientific). Samples were vortexed before centrifugation for 10 min at 21,300g at 20°C. Clear supernatants were diluted 1:10 with LC-MS grade water and transferred to fresh autosampler tubes with conical glass inserts (Chromatographie Zubehoer Trott) and analyzed using a Vanquish UHPLC (Thermo Fisher Scientific) connected to a Q-Exactive HF (Thermo Fisher Scientific).
For the analysis, 1 μl of the derivatized sample were injected onto a 100 × 2.1 mm HSS T3 UPLC column (Waters). The flow rate was set to 400 μl/min using a buffer system consisting of buffer A [10 mM ammonium formate (Sigma-Aldrich) and 0.15% formic acid (Sigma-Aldrich) in LC-MS grade water] and buffer B (acetonitrile, Optima-grade, Fisher Scientific). The LC gradient was 0% buffer B at 0 min, 0 to 15% buffer B at 0 to 0.1 min, 15 to 17% buffer B at 0.1 to 0.5 min, 17 to 55% buffer B at 0.5 to 7 min, 55 to 70% buffer B at 7 to 7.5 min, 70 to 100% buffer B at 7.5 to 9 min, 100% buffer B at 9 to 10 min, 100 to 0% buffer B at 10 to 10.1 min, and 0% buffer B at 10.1 to 15 min. The mass spectrometer was operating in positive-ionization mode monitoring the mass range, mass/charge ratio of 50 to 750. The heated electrospray ionization (ESI) source settings of the mass spectrometer were as follows: spray voltage of 3.5 kV, capillary temperature of 250°C, sheath gas flow of 60 arbitrary units (AU), and auxiliary gas flow of 20 AU at a temperature of 250°C. The S-lens was set to a value of 60 AU.
Data analysis was performed using the TraceFinder software (version 4.2, Thermo Fisher Scientific). Identity of each compound was validated by authentic reference compounds, which were analyzed independently. Peak areas were analyzed using extracted ion chromatogram (XIC) of compound-specific [M + nBz + H]+, where n corresponds to the number of amine moieties, which can be derivatized with a benzoyl chloride (Bz). XIC peaks were extracted with a mass accuracy (<5 parts per million) and a retention time tolerance of 0.2 min.
Anion-exchange chromatography MS of the analysis of tricarboxylic acid cycle and glycolysis metabolites
Anion-exchange chromatography was performed simultaneously to the LC-MS analysis. First, 50 μl of the resuspended sample was diluted 1:5 with LC-MS grade water and analyzed using a Dionex ion chromatography system (ICS-5000, Thermo Fisher Scientific). The applied protocol was adopted from (70). Briefly, 10 μl of polar metabolite extract was injected in full-loop mode using an overfill factor of 3, onto a Dionex IonPac AS11-HC column (2 mm by 250 mm, 4-μm particle size, Thermo Fisher Scientific) equipped with a Dionex IonPac AS11-HC guard column (2 mm by 50 mm, 4 μm, Thermo Fisher Scientific). The column temperature was held at 30°C, while the autosampler was set to 6°C. A potassium hydroxide gradient was generated by the eluent generator using a potassium hydroxide cartridge that was supplied with deionized water. The metabolite separation was carried at a flow rate of 380 μl/min, applying the following gradient: 0 to 5 min, 10 to 25 mM KOH; 5 to 21 min, 25 to 35 mM KOH; 21 to 25 min, 35 to 100 mM KOH, 25 to 28 min, 100 mM KOH; and 28 to 32 min, 100 to 10 mM KOH. The column was re-equilibrated at 10 mM for 6 min. The eluting metabolites were detected in negative ion mode using ESI MRM (multireaction monitoring) on a Xevo TQ (Waters) triple quadrupole mass spectrometer applying the following settings: capillary voltage of 1.5 kV, desolvation temperature of 550°C, desolvation gas flow of 800 liters/hour, and collision cell gas flow of 0.15 ml/min. All peaks were validated using two MRM transitions, one for quantification of the compound, while the second ion was used for qualification of the identity of the compound. Data analysis and peak integration were performed using the TargetLynx Software (Waters).
Analysis of nuclear translocation of TFEB
Images for assessment of nuclear translocation of TFEB-GFP in paraformaldehyde-fixed N2a cells were acquired using the VisiScope CSU-W1 spinning disk confocal microscope and the VisiView Software (Visitron Systems GmbH). Settings for laser and detector were maintained constant for the acquisition of each image. For analysis, at least three images were acquired using the 20× objective. For quantification of nuclear TFEB- GFP translocation, GFP intensity was determined in ImageJ by manually drawing a border around randomly selected, 4′,6-diamidino-2-phenylindole–positive nuclei of N2a cells with a GFP signal.
Statistical analysis
The data presented are shown as means ± SEM, and samples sizes are indicated in the figure legends. All data were analyzed by the commercially available software SPSS v17.0 and GraphPad v8.0. The unpaired Student’s t test was used when two groups were compared. For four-group comparisons, two-way analysis of variance (ANOVA) was performed, followed by Tukey’s or Dunnett’s multiple comparisons test, as appropriate. P values of less than 0.05 were considered statistically significant.
Acknowledgments
We thank C. Kühne, D. Harbich and B. Schmid (Max Planck Institute of Psychiatry, Munich, Germany) for excellent technical assistant and support. We thank J. Deussing and the scientific core unit Genetically Engineered Mouse Models for providing technical support and guidance. We thank the Microscopy Core Facility of the Medical Faculty at the University of Bonn for providing support and instrumentation funded by the Deutsche Forschungsgemeinschaft, project number: 388169927.
Funding: This work was supported by the “OptiMD” grant of the Federal Ministry of Education and Research (01EE1401D to M.V.S.), the BioM M4 award “PROCERA” of the Bavarian State Ministry (to M.V.S.), the “GUTMOM” grant of the Federal Ministry of Education and Research (01EA1805, to M.V.S.), and the “Kids2Health” grant of the Federal Ministry of Education and Research (01GL1743C, to M.V.S.).
Author contributions: A.S.H, G.B., M.V.S., and N.C.G. conceived the project and designed the experiments. A.S.H and L.M.B. managed the mouse lines and genotyping. A.S.H., M.L.P., and L.M.B. performed animal experiments and surgeries. K.H., T.B., and N.C.G. performed protein work. P.G. and T.B. performed and analyzed metabolomics experiments. M.D.A, J.N., and K.-W. S. performed catecholamine extraction. A.S.H. wrote the initial version of the manuscript. M.V.S., A.C., and N.C.G. supervised the research, and all authors revised the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials and in the source data table. All data needed to interpret the metabolomics analyses are included in the source data file. Raw files of MS measurements are deposited at Zenodo and can be accessed using the following DOIs: For Bz measurements, 10.5281/zenodo.5772429; and for IC measurements, 10.5281/zenodo.5775917. Sample IDs can be inferred from the source data file “MS_SAMPLE_IDs_metabolomics.
Supplementary Materials
This PDF file includes:
Figs. S1 to S10
Other Supplementary Material for this manuscript includes the following:
Data S1
REFERENCES AND NOTES
- 1.Morton G. J., Meek T. H., Schwartz M. W., Neurobiology of food intake in health and disease. Nat. Rev. Neurosci. 15, 367–378 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galluzzi L., Bravo-San Pedro J. M., Levine B., Green D. R., Kroemer G., Pharmacological modulation of autophagy: Therapeutic potential and persisting obstacles. Nat. Rev. Drug Discov. 16, 487–511 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Y., Sowers J. R., Ren J., Targeting autophagy in obesity: From pathophysiology to management. Nat. Rev. Endocrinol. 14, 356–376 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Rabinowitz J. D., White E., Autophagy and metabolism. Science 330, 1344–1348 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levine B., Kroemer G., Biological functions of autophagy genes: A disease perspective. Cell 176, 11–42 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Potes Y., de Luxán-Delgado B., Rodriguez-González S., Guimarães M. R. M., Solano J. J., Fernández-Fernández M., Bermúdez M., Boga J. A., Vega-Naredo I., Coto-Montes A., Overweight in elderly people induces impaired autophagy in skeletal muscle. Free Radic. Biol. Med. 110, 31–41 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Mizunoe Y., Sudo Y., Okita N., Hiraoka H., Mikami K., Narahara T., Negishi A., Yoshida M., Higashibata R., Watanabe S., Kaneko H., Natori D., Furuichi T., Yasukawa H., Kobayashi M., Higami Y., Involvement of lysosomal dysfunction in autophagosome accumulation and early pathologies in adipose tissue of obese mice. Autophagy 13, 642–653 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inoki K., Kim J., Guan K.-L., AMPK and mTOR in cellular energy homeostasis and drug targets. Annu. Rev. Pharmacol. Toxicol. 52, 381–400 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Mihaylova M. M., Shaw R. J., The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat. Cell Biol. 13, 1016–1023 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim J., Kundu M., Viollet B., Guan K. L., AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 13, 132–141 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bakula D., Müller A. J., Zuleger T., Takacs Z., Franz-Wachtel M., Thost A. K., Brigger D., Tschan M. P., Frickey T., Robenek H., Macek B., Proikas-Cezanne T., WIPI3 and WIPI4 β-propellers are scaffolds for LKB1-AMPK-TSC signalling circuits in the control of autophagy. Nat. Commun. 8, 15637 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaushik S., Arias E., Kwon H., Lopez N. M., Athonvarangkul D., Sahu S., Schwartz G. J., Pessin J. E., Singh R., Loss of autophagy in hypothalamic POMC neurons impairs lipolysis. EMBO Rep. 13, 258–265 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaushik S., Rodriguez-Navarro J. A., Arias E., Kiffin R., Sahu S., Schwartz G. J., Cuervo A. M., Singh R., Autophagy in hypothalamic AgRP neurons regulates food intake and energy balance. Cell Metab. 14, 173–183 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riggs D. L., Roberts P. J., Chirillo S. C., Cheung-Flynn J., Prapapanich V., Ratajczak T., Gaber R., Picard D., Smith D. F., The Hsp90-binding peptidylprolyl isomerase FKBP52 potentiates glucocorticoid signaling in vivo. EMBO J. 22, 1158–1167 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wochnik G. M., Rüegg J., Abel G. A., Schmidt U., Holsboer F., Rein T., FK506-binding proteins 51 and 52 differentially regulate dynein interaction and nuclear translocation of the glucocorticoid receptor in mammalian cells. J. Biol. Chem. 280, 4609–4616 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Taipale M., Tucker G., Peng J., Krykbaeva I., Lin Z. Y., Larsen B., Choi H., Berger B., Gingras A. C., Lindquist S., A quantitative chaperone interaction network reveals the architecture of cellular protein homeostasis pathways. Cell 158, 434–448 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt M. V., Paez-Pereda M., Holsboer F., Hausch F., The prospect of FKBP51 as a drug target. ChemMedChem 7, 1351–1359 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Balsevich G., Häusl A. S., Meyer C. W., Karamihalev S., Feng X., Pöhlmann M. L., Dournes C., Uribe-Marino A., Santarelli S., Labermaier C., Hafner K., Mao T., Breitsamer M., Theodoropoulou M., Namendorf C., Uhr M., Paez-Pereda M., Winter G., Hausch F., Chen A., Tschöp M. H., Rein T., Gassen N. C., Schmidt M. V., Stress- responsive FKBP51 regulates AKT2-AS160 signaling and metabolic function. Nat. Commun. 8, 1725 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stechschulte L. A., Qiu B., Warrier M., Hinds T. D., Zhang M., Gu H., Xu Y., Khuder S. S., Russo L., Najjar S. M., Lecka-Czernik B., Yong W., Sanchez E. R., FKBP51 null mice are resistant to diet-induced obesity and the PPARγ agonist rosiglitazone. Endocrinology 157, 3888–3900 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pereira M. J., Palming J., Svensson M. K., Rizell M., Dalenbäck J., Hammar M., Fall T., Sidibeh C. O., Svensson A., Eriksson J. W., FKBP5 expression in human adipose tissue increases following dexamethasone exposure and is associated with insulin resistance. Metabolism 63, 1198–1208 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Sidibeh C. O., Pereira M. J., Abalo X. M., Boersma G. J., Skrtic S., Lundkvist P., Katsogiannos P., Hausch F., Castillejo-López C., Eriksson J. W., FKBP5 expression in human adipose tissue: Potential role in glucose and lipid metabolism, adipogenesis and type 2 diabetes. Endocrine 62, 116–128 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Häusl A. S., Balsevich G., Gassen N. C., Schmidt M. V., Focus on FKBP51: A molecular link between stress and metabolic disorders. Mol. Metab. 29, 170–181 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pei H., Li L., Fridley B. L., Jenkins G. D., Kalari K. R., Lingle W., Petersen G., Lou Z., Wang L., FKBP51 affects cancer cell response to chemotherapy by negatively regulating Akt. Cancer Cell 16, 259–266 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang R. C., Wei Y., An Z., Zou Z., Xiao G., Bhagat G., White M., Reichelt J., Levine B., Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science 338, 956–959 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gassen N. C., Hartmann J., Schmidt M. V., Rein T., FKBP5/FKBP51 enhances autophagy to synergize with antidepressant action. Autophagy 11, 578–580 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gassen N. C., Niemeyer D., Muth D., Corman V. M., Martinelli S., Gassen A., Hafner K., Papies J., Mösbauer K., Zellner A., Zannas A. S., Herrmann A., Holsboer F., Brack-Werner R., Boshart M., Müller-Myhsok B., Drosten C., Müller M. A., Rein T., SKP2 attenuates autophagy through Beclin1-ubiquitination and its inhibition reduces MERS-Coronavirus infection. Nat. Commun. 10, 5770 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M., Tanaka K., Cuervo A. M., Czaja M. J., Autophagy regulates lipid metabolism. Nature 458, 1131–1135 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toneatto J., Guber S., Charo N. L., Susperreguy S., Schwartz J., Galigniana M. D., Piwien-Pilipuk G., Dynamic mitochondrial-nuclear redistribution of the immunophilin FKBP51 is regulated by the PKA signaling pathway to control gene expression during adipocyte differentiation. J. Cell Sci. 126, 5357–5368 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y., Goldman S., Baerga R., Zhao Y., Komatsu M., Jin S., Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc. Natl. Acad. Sci. U.S.A. 106, 19860–19865 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang L., Isoda F., Yen K., Kleopoulos S. P., Janssen W., Fan X., Mastaitis J., Dunn-Meynell A., Levin B., McCrimmon R., Sherwin R., Musatov S., Mobbs C. V., Hypothalamic Fkbp51 is induced by fasting, and elevated hypothalamic expression promotes obese phenotypes. AJP Endocrinol. Metab. 302, E987–E991 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scharf S. H., Liebl C., Binder E. B., Schmidt M. V., Müller M. B., Expression and regulation of the Fkbp5 gene in the adult mouse brain. PLOS ONE 6, e16883 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolfson R. L., Sabatini D. M., The dawn of the age of amino acid sensors for the mTORC1 pathway. Cell Metab. 26, 301–309 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Son S. M., Park S. J., Lee H., Siddiqi F., Lee J. E., Menzies F. M., Rubinsztein D. C., Leucine signals to mTORC1 via its metabolite acetyl-coenzyme A. Cell Metab. 29, 192–201.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meng D., Yang Q., Wang H., Melick C. H., Navlani R., Frank A. R., Jewell J. L., Glutamine and asparagine activate mTORC1 independently of Rag GTPases. J. Biol. Chem. 295, 2890–2899 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klionsky D. J., Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12, 1–222 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arif A., Jia J., Willard B., Li X., Fox P. L., Multisite phosphorylation of S6K1 directs a kinase phospho-code that determines substrate selection. Mol. Cell. 73, 446–457.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim J., Kim Y. C., Fang C., Russell R. C., Kim J. H., Fan W., Liu R., Zhong Q., Guan K. L., Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell 152, 290–303 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shackelford D. B., Shaw R. J., The LKB1-AMPK pathway: Metabolism and growth control in tumour suppression. Nat. Rev. Cancer 9, 563–575 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tian W., Alsaadi R., Guo Z., Kalinina A., Carrier M., Tremblay M. E., Lacoste B., Lagace D., Russell R. C., An antibody for analysis of autophagy induction. Nat. Methods 17, 232–239 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Zhang H., Alsaleh G., Feltham J., Sun Y., Napolitano G., Riffelmacher T., Charles P., Frau L., Hublitz P., Yu Z., Mohammed S., Ballabio A., Balabanov S., Mellor J., Simon A. K., Polyamines control eIF5A hypusination, TFEB translation, and autophagy to reverse B cell senescence. Mol. Cell. 76, 110–125.e9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wan W., You Z., Zhou L., Xu Y., Peng C., Zhou T., Yi C., Shi Y., Liu W., mTORC1-regulated and HUWE1-mediated WIPI2 degradation controls autophagy Flux. Mol. Cell. 72, 303–315.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Liu G. Y., Sabatini D. M., mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. 21, 183–203 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Broberger C., Brain regulation of food intake and appetite: Molecules and networks. J. Intern. Med. 258, 301–327 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Brodie B. B., Costa E., Dlabac A., Neff N. H., Smookler H. H., Application of steady state kinetics to the estimation of synthesis rate and turnover time of tissue catecholamines. J. Pharmacol. Exp. Ther. 154, 493–498 (1966). [PubMed] [Google Scholar]
- 45.Joly-Amado A., Denis R. G. P., Castel J., Lacombe A., Cansell C., Rouch C., Kassis N., Dairou J., Cani P. D., Ventura-Clapier R., Prola A., Flamment M., Foufelle F., Magnan C., Luquet S., Hypothalamic AgRP-neurons control peripheral substrate utilization and nutrient partitioning. EMBO J. 31, 4276–4288 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sperringer J. E., Addington A., Hutson S. M., Branched-chain amino acids and brain metabolism. Neurochem. Res. 42, 1697–1709 (2017). [DOI] [PubMed] [Google Scholar]
- 47.J. D. Fernstrom, Journal of Nutrition (American Institute of Nutrition, 2005; https://academic.oup.com/jn/article/135/6/1539S/4663842), vol. 135, pp. 1539S–1546S.
- 48.Newgard C. B., An J., Bain J. R., Muehlbauer M. J., Stevens R. D., Lien L. F., Haqq A. M., Shah S. H., Arlotto M., Slentz C. A., Rochon J., Gallup D., Ilkayeva O., Wenner B. R., Yancy W. S., Eisenson H., Musante G., Surwit R. S., Millington D. S., Butler M. D., Svetkey L. P., A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 9, 311–326 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang T. J., Larson M. G., Vasan R. S., Cheng S., Rhee E. P., McCabe E., Lewis G. D., Fox C. S., Jacques P. F., Fernandez C., O’Donnell C. J., Carr S. A., Mootha V. K., Florez J. C., Souza A., Melander O., Clish C. B., Gerszten R. E., Metabolite profiles and the risk of developing diabetes. Nat. Med. 17, 448–453 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoon M. S., The emerging role of branched-chain amino acids in insulin resistance and metabolism. Nutrients. 8, 405 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheng Y., Zhang Q., Meng Q., Xia T., Huang Z., Wang C., Liu B., Chen S., Xiao F., Du Y., Guo F., Leucine deprivation stimulates fat loss via increasing CRH expression in the hypothalamus and activating the sympathetic nervous system. Mol. Endocrinol. 25, 1624–1635 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mariño G., Pietrocola F., Eisenberg T., Kong Y., Malik S. A., Andryushkova A., Schroeder S., Pendl T., Harger A., Niso-Santano M., Zamzami N., Scoazec M., Durand S., Enot D. P., Fernández Á. F., Martins I., Kepp O., Senovilla L., Bauvy C., Morselli E., Vacchelli E., Bennetzen M., Magnes C., Sinner F., Pieber T., López-Otín C., Maiuri M. C., Codogno P., Andersen J. S., Hill J. A., Madeo F., Kroemer G., Regulation of autophagy By cytosolic acetyl-coenzyme A. Mol. Cell 53, 710–725 (2014). [DOI] [PubMed] [Google Scholar]
- 53.Son S. M., Park S. J., Stamatakou E., Vicinanza M., Menzies F. M., Rubinsztein D. C., Leucine regulates autophagy via acetylation of the mTORC1 component raptor. Nat. Commun. 11, 3148 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pietrocola F., Lachkar S., Enot D. P., Niso-Santano M., Bravo-San Pedro J. M., Sica V., Izzo V., Maiuri M. C., Madeo F., Mariño G., Kroemer G., Spermidine induces autophagy by inhibiting the acetyltransferase EP300. Cell Death Differ. 22, 509–516 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sacitharan P. K., Lwin S., Gharios G. B., Edwards J. R., Spermidine restoresdysregulated autophagy and polyamine synthesis in aged and osteoarthritic chondrocytes via EP300. Exp. Mol. Med. 50, 1–10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hardie D. G., AMP-activated/SNF1 protein kinases: Conserved guardians of cellular energy. Nat. Rev. Mol. Cell Biol. 8, 774–785 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Stechschulte L. A., Hinds T. D., Khuder S. S., Shou W., Najjar S. M., Sanchez E. R., FKBP51 controls cellular adipogenesis through p38 kinase-mediated phosphorylation of GRα and PPARγ. Mol. Endocrinol. 28, 1265–1275 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meng Q., Cai D., Defective hypothalamic autophagy directs the central pathogenesis of obesity via the IκB kinase β(IKKβ)/NF-κB pathway. J. Biol. Chem. 286, 32324–32332 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Myers M. G., Olson D. P., Central nervous system control of metabolism. Nature 491, 357–363 (2012). [DOI] [PubMed] [Google Scholar]
- 60.Um S. H., Frigerio F., Watanabe M., Picard F., Joaquin M., Sticker M., Fumagalli S., Allegrini P. R., Kozma S. C., Auwerx J., Thomas G., Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature 431, 200–205 (2004). [DOI] [PubMed] [Google Scholar]
- 61.Khamzina L., Veilleux A., Bergeron S., Marette A., Increased activation of the mammalian target of rapamycin pathway in liver and skeletal muscle of obese rats: Possible involvement in obesity-linked insulin resistance. Endocrinology 146, 1473–1481 (2005). [DOI] [PubMed] [Google Scholar]
- 62.Cota D., Proulx K., Smith K. A. B., Kozma S. C., Thomas G., Woods S. C., Seeley R. J., Hypothalamic mTOR signaling regulates food intake. Science 312, 927–930 (2006). [DOI] [PubMed] [Google Scholar]
- 63.Blouet C., Ono H., Schwartz G. J., Mediobasal hypothalamic p70 S6 kinase 1 modulates the control of energy homeostasis. Cell Metab. 8, 459–467 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dagon Y., Hur E., Zheng B., Wellenstein K., Cantley L. C., Kahn B. B., P70S6 kinase phosphorylates AMPK on serine 491 to mediate leptin’s effect on food intake. Cell Metab. 16, 104–112 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Häusl A., Hartmann J., Pöhlmann M., Brix L., Lopez J.-P., Brivio E., Engelhardt C., Roeh S., Rudolph L., Stoffel R., Hafner K., Goss H., Reul J., Deussing J., Ressler K., Gassen N., Chen A., Schmidt M., The co-chaperone Fkbp5 shapes the acute stress response in the paraventricular nucleus of the hypothalamus. bioRxiv, 824664 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schmidt M. V., Schulke J.-P., Liebl C., Stiess M., Avrabos C., Bock J., Wochnik G. M., Davies H. A., Zimmermann N., Scharf S. H., Trumbach D., Wurst W., Zieglgansberger W., Turck C., Holsboer F., Stewart M. G., Bradke F., Eder M., Muller M. B., Rein T., Tumor suppressor down-regulated in renal cell carcinoma 1 (DRR1) is a stress-induced actin bundling factor that modulates synaptic efficacy and cognition. Proc. Natl. Acad. Sci. 108, 17213–17218 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nagler J., Schriever S. C., De Angelis M., Pfluger P. T., Schramm K. W., Comprehensive analysis of nine monoamines and metabolites in small amounts of peripheral murine (C57Bl/6 J) tissues. Biomed. Chromatogr. 32, e4151 (2018). [DOI] [PubMed] [Google Scholar]
- 68.Martinelli S., Anderzhanova E. A., Wiechmann S., Dethloff F., Weckmann K., Bajaj T., Hartmann J., Hafner K., Pöhlmann M. L., Jollans L., Maccarrone G., Hausch F., Turck C. W., Philipsen A., Schmidt M. V., Kuster B., Gassen N. C., Stress-primed secretory autophagy drives extracellular BDNF maturation. bioRxiv 2020, 2020.05.13.090514 (2020). [Google Scholar]
- 69.Wong J. M. T., Malec P. A., Mabrouk O. S., Ro J., Dus M., Kennedy R. T., Benzoyl chloride derivatization with liquid chromatography-mass spectrometry for targeted metabolomics of neurochemicals in biological samples. J. Chromatogr. A 1446, 78–90 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schwaiger M., Rampler E., Hermann G., Miklos W., Berger W., Koellensperger G., Anion-exchange chromatography coupled to high-resolution mass spectrometry: A powerful tool for merging targeted and non-targeted metabolomics. Anal. Chem. 89, 7667–7674 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S10
Data S1