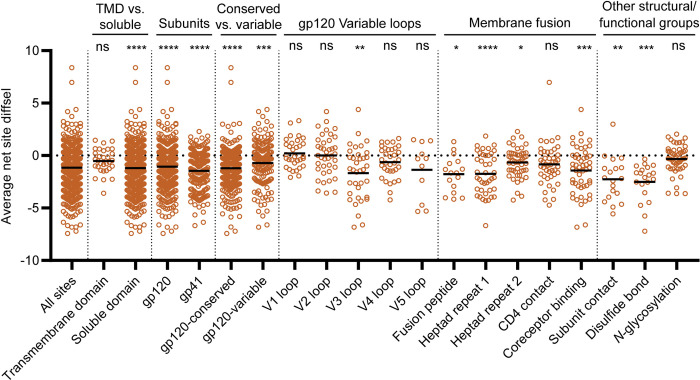
Fig 5. Impact of XBP1s induction on mutational tolerance varies across Env structural elements.
Average net site differential selection (diffsel) for the +XBP1s endoplasmic reticulum (ER) proteostasis environment normalized to the basal ER proteostasis environment, where the means of the distributions are indicated by black horizontal lines. Sites are sorted by transmembrane domain (TMD) versus soluble, subunits, conserved versus variable regions of gp120, the 5 variable loops of gp120, regions important for membrane fusion, and other structural/functional groups. For TMD versus soluble, all sites that do not belong to the TMD were categorized as “soluble.” For conserved versus variable, the sites that belong to the 5 variable loops of gp120 were categorized as “gp120–variable,” and the sites that are not included in the 5 variable loops were categorized as “gp120–conserved.” Significance of deviation from null (net site diffsel = 0, no selection) was tested using a 1-sample t test. The derived p-values were Bonferroni-corrected for 20 tests; *p-value < 0.05, **p-value < 0.01, ***p-value < 0.001, ****p-value < 0.0001; ns, not significant. Diffsel values are provided at https://github.com/yoon-jimin/2021_HIV_Env_DMS. Assignment of structural regions is provided in S2 Table.