Fig 6. Diverse functional elements of Env respond differently to XBP1s induction.
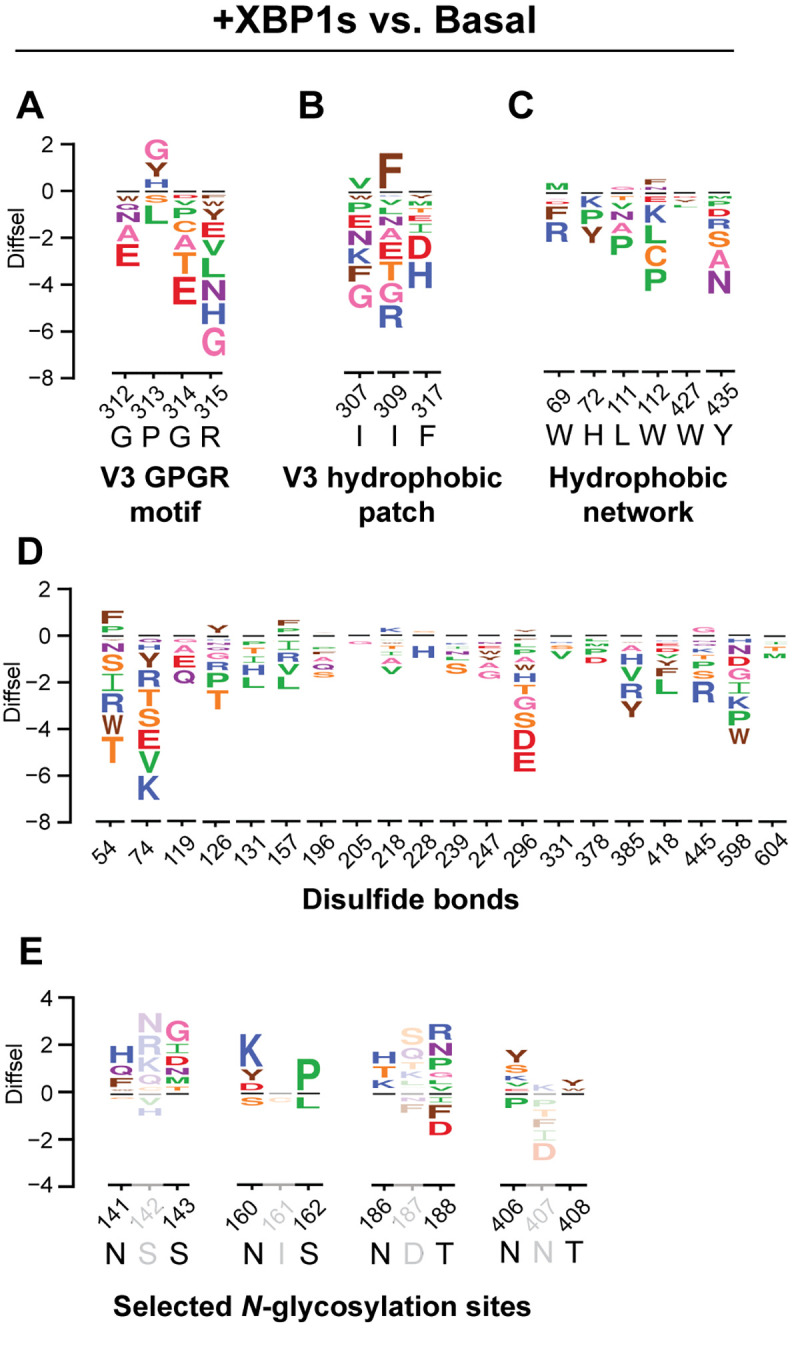
Selected sequence logo plots for the +XBP1s endoplasmic reticulum (ER) proteostasis environment normalized to the basal ER proteostasis environment for (A) the conserved GPGR motif of the V3 loop, (B) the hydrophobic patch of the V3 loop, (C) the hydrophobic network of gp120 (important for CD4 binding), (D) cysteine residues participating in disulfide bonds, and (E) selected N-glycosylation sequons (N-X-S/T) that exhibited positive net site differential selection (diffsel) in all 3 remodeled proteostasis environments. The height of the amino acid abbreviation corresponds to the magnitude of diffsel. The numbers and letters below the logos indicate the Env site in HXB2 numbering and the wild-type amino acid for that site, respectively. Only variants that were present in all 3 pre-selection viral libraries and exhibited diffsel in the same direction across the biological triplicates are plotted. All logo plots were generated on the same scale. Diffsel values are provided at https://github.com/yoon-jimin/2021_HIV_Env_DMS. Assignments of functional regions are provided in S2 Table.
