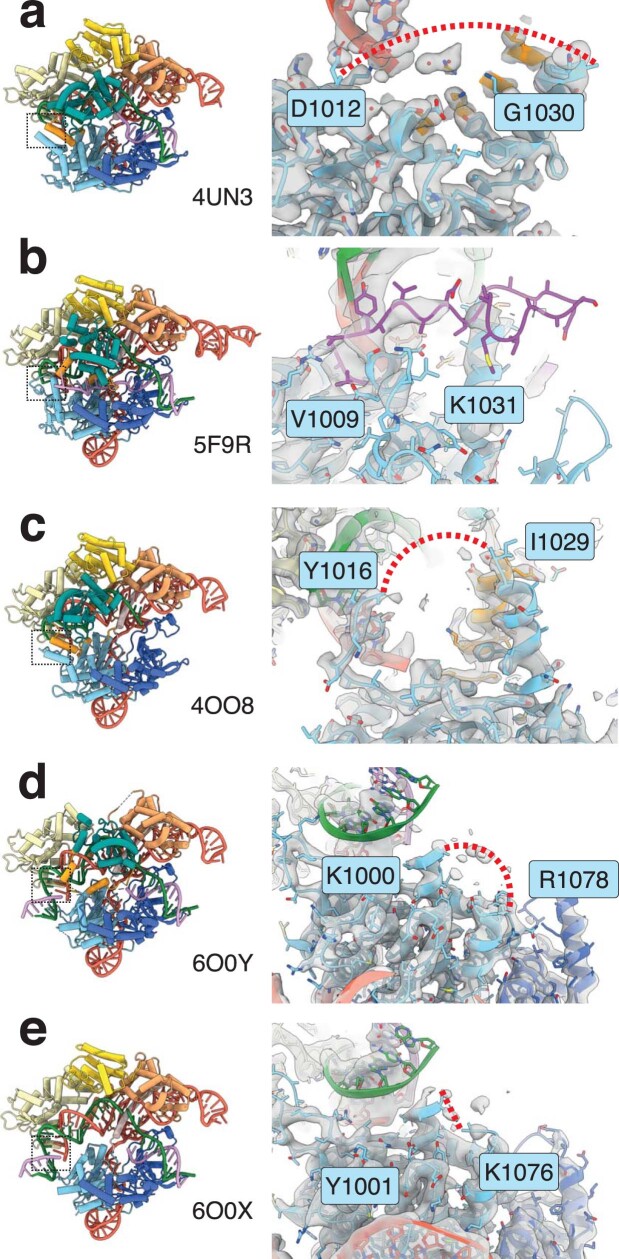
Extended Data Fig. 6. RuvC loop in on-target SpCas9 structures.
a, On-target inactive Cas9 bound to dsDNA (PDB 4UN3)15. RuvC loop is missing between 1013–1029. b, On-target inactive (primed – HNH rearranged and adjacent to target strand scissile phosphate) Cas9 bound to dsDNA (PDB 5F9R)14. RuvC loop has been built primarily as alanine ‘stub’ residues, but electron density is very poor and diffuse for this region. c, On-target inactive Cas9 bound to dsDNA (PDB 4OO8)43. RuvC loop is missing between 1017–1028. d, On-target active Cas9 bound to dsDNA in postcatalysis state18. RuvC loop is missing between 1001–1077. e, On-target active Cas9 bound to dsDNA in product state18. RuvC loop is missing between 1000–1075. In a–c, electron density is displayed as a grey surface, and in d, e cryo-EM density is shown as a grey surface. In all structures, missing residues are depicted as a red dashed line with the RuvC loop in b shown as magenta. Position of RuvC loop is denoted by a black dashed box in the left panel for each model.